 |
D-Lib Magazine
September/October 2013
Volume 19, Number 9/10
Table of Contents
Extraction of References Using Layout and Formatting Information from Scientific Articles
Roman Kern
Knowledge Technologies Institute, Graz University of Technology, Graz, Austria
rkern@tugraz.at
Stefan Klampfl
Know-Center, Graz, Austria
sklampfl@know-center.at
doi:10.1045/september2013-kern
Printer-friendly Version
Abstract
The automatic extraction of reference meta-data is an important requirement for the efficient management of collections of scientific literature. An existing powerful state-of-the-art system for extracting references from a scientific article is ParsCit; however, it requires the input document to be converted into plain text, thereby ignoring most of the formatting and layout information. In this paper, we quantify the contribution of this additional information to the reference extraction performance by an improved preprocessing using the information contained in PDF files and retraining sequence classifiers on an enhanced feature set. We found that the detection of columns, reading order, and decorations, as well as the inclusion of layout information improves the retrieval of reference strings, and the classification of reference token types can be improved using additional font information. These results emphasize the importance of layout and formatting information for the extraction of meta-data from scientific articles.
Keywords: Content Analysis and Indexing, Meta-data Extraction, Natural Language Processing, Text Analysis, Supervised Machine Learning
1. Introduction
Scientific articles usually conclude with a section that acknowledges relevant and related work in the form of a list of citations or references. Researchers often use this information to search for literature that is relevant for their own work or for a specific field of research. With the growth of the global volume of scientific literature reaching unprecedented levels, there is an increasing demand for automated processing systems that support researchers in this task, e.g., through the creation of citation networks such as CiteSeer[5], which allow a convenient navigation through a specific field of research forward and backward in time.
The automated extraction of reference meta-data requires the detection of the reference section within a paper, the segmentation of individual reference strings, and the labelling of single tokens within each string as to which field they belong (e.g., author, title, year, journal). These are challenging tasks given the variety of different journal layouts and formatting of reference strings. Several approaches have been proposed using unsupervised methods [3] and template matching [7]; however, the most promising method has been the supervised classification of sequences, e.g., through Hidden Markov Models (HMMs) [14] or Conditional Random Fields (CRFs) [12]. The sequence classification approach has also been prominently used for the extraction of other types of meta-data from scientific articles [11, 6, 8]. ParsCit [4] is an existing state-of-the-art reference extraction system that uses heuristics to detect and segment references within a scientific article, and CRFs to assign labels to the tokens within each reference string. However, this system assumes the input article was provided in raw text format, thereby ignoring most of the formatting and layout information, such as columns, line indentation, or font style. As these features certainly help humans to read and understand references, their inclusion should intuitively improve the performance of the reference extraction.
In this paper we quantify the contribution of formatting and layout information provided by PDF files, the most common format of scientific literature today, to the reference extraction performance. In particular, we incorporate information about columns, line gaps and indentations, and font styles into both the extraction of reference strings and the classification of their tokens. We retrained ParsCit on a representative document set using both the original feature set in [4] and a layout-enhanced feature set. We also compare the performance values obtained by different sequence classifiers and by the out-of-the-box ParsCit system available for download.
This paper is structured as follows. In section 2 we devise the individual steps of our reference extraction algorithm and highlight the enhancements we made with respect to the original ParsCit system. In section 3 we separately evaluate the individual stages of our algorithm, reference segmentation and token classification. We discuss the results in section 4 before we conclude with an outlook to future work in section 5.
2. Algorithm
In this section we describe the individual steps of our reference extraction algorithm. It takes a scientific article in PDF format as input and produces a list of references with categorized tokens.
2.1 Reference Line Extraction
The first step is to find the lines containing the references within an article. The starting point for the reference line extraction is a set of contiguous text blocks of the input PDF file. These text blocks are provided by an open source tool that builds upon the output of the PDFBox library. The resulting physical structure consists of a list of text blocks hierarchically organized into lines, words, and characters, as well as the reading order of blocks within the document [1]. Most importantly, this block structure effectively provides a segmentation of the text into single columns, a fact that is particularly helpful for reference extraction.
The procedure for extracting the reference lines is similar to the preprocessing step of the original ParsCit algorithm [4]. We iterate over all blocks in the reading order and use a regular expression to find the reference headers, which we assume to be one of "References", "Bibliography", "References and Notes", "Literature cited", and common variations of those strings (e.g., uppercase variants). Additionally we allow five extra characters in the heading, in case there is a section numbering or a related decoration. Then we collect all lines until we encounter either another section heading, starting with "Acknowledgement", "Autobiographical", "Table", "Appendix", "Exhibit", "Annex", "Fig", or "Notes", or the end of the document.
Compared with the original ParsCit system, we incorporated additional layout information into the reference line extraction in three ways. First, column information is implicitly provided through the consideration of text blocks. Second, we ignored the content of decoration blocks (headers and footers consisting of page numbers, authors, or journal names), which we computed by associating blocks across neighboring pages based on their content and geometrical position [10]. Finally, we ignored all lines following a vertical gap that is larger than the average gap size plus two times the standard deviation of gap sizes. This criterion has been introduced as the block of references is often followed by footnotes, copyright information or other types of text which is visually separated from the references by a bigger gap.
The output of the references line extraction step is a collection of lines containing the article's references.
2.2 Reference Segmentation
After the reference lines of the article have been collected, the next step is the segmentation of these lines into individual reference strings. The input to this step is a collection of reference lines and the output is a list of extracted reference strings.
The original ParsCit algorithm distinguishes between three cases: 1) reference strings are marked with square or round brackets (e.g., "[1]" or "(1)"), 2) strings are marked with naked numbers, and 3) strings are unmarked. For cases 1 and 2 the most common marker type is found via regular expressions, and the marker is also used to segment the references. For case 3, heuristics such as the length of lines or ending punctuations are used to decide where a new reference string starts.
In contrast with ParsCit, our implementation makes use of the layout information within the PDF. The layout-based splitting comes into play in case 3, when there are no known reference markers. The basis of our reference splitting algorithm is the observation that for many reference styles the first line of a reference has a negative indentation. Thus, our algorithm looks for lines which visually stick out from the rest of the lines, assuming that the first lines will be the minority class.
We inspect each text block containing reference lines. If a block contains just a single line, this line is assumed to be an artifact of the PDF extraction process and is completely ignored. For blocks with more than two lines we cluster the lines using a simple version of the k-means clustering algorithm. The sole feature we use for this is the minimal x-coordinate of a line's bounding box. We set the number of clusters to 2 and initialize the two centroids with the minimal and maximal value of the feature. Then we assign each line to that centroid which is closer to the line's x-coordinate. We stop after a single iteration and update the centroids with the mean of the assigned features. At this stage all lines are assigned to one of the two clusters. Only if two conditions are met is the layout-based splitting applied: The minimum cluster must contain fewer lines than the maximum cluster and the centroids differ by at least 0.05 ∗ maxLineWidth. If this is the case, all lines from the minimum cluster are considered to be the first line of a new reference at which the reference lines are split into individual reference strings.
2.3 Reference Preprocessing
The task performed by the reference preprocessing step is to clean the text of the references before the token classification is applied. The preprocessing consists of two parts, dehyphenation and normalization.
In the first part we resolve hyphenations by removing hyphens "-" and concatenating the split word parts if they are the result of a proper English hyphenation. For each line that ends with a hyphen we apply hyphenation on the concatenated word using a list of hyphenation patterns taken from the TEX distribution, and if the line split occurs at one of the proposed split points we resolve the hyphenation.
In the normalization part, we align the pages information to the form "<number>--<number>", even if there are multiple tokens or different dash characters.
These preprocessing steps are not part of the original ParsCit approach.
2.4 Reference Token Classification
The final step is the categorization of the individual tokens of the extracted and preprocessed reference strings. We used the following token types: authorGivenName, authorSurname, authorOther, editor, title, date, publisher, issueTitle, bookTitle, pages, location, conference, source, volume, edition, issue, url, note, and other. These classes differ slightly from the original ParsCit system. In particular, the author class is split into three classes: authorGivenName, authorSurname and authorOthers, where the latter is used for intermediate tokens in the author substring, such as "and".
At the core of the reference extraction process lies a supervised machine learning algorithm that takes sequence information into account. We use two different sequence classification algorithms: one is a conditional random field (CRF) [9], which has also been used in the original ParsCit system. As implementation we use the freely available crfsuite software. The other classification algorithm is provided by the open-source library OpenNLP, the Maximum Entropy classifier [2]. In order to integrate sequence information, we follow a Beam Search approach [13], which takes the classification decision of a number of preceding instances into account (here: 4).
We use all the original features from the original ParsCit approach (see section 2 in [4] for the complete list). For Beam Search we generate 2 additional features: seenLabels, the set of all labels that occurred previously in the current reference string, and finishedLabels, the set of all labels that occurred previously and were followed by at least one other label. In addition, we incorporate layout and formatting information by a set of binary features specifying whether the font of the tokens inside a sliding window from -2 to +2 tokens is equal to the font of the current token. Two fonts are considered equal if they share the same font name, the same font size, and the same binary attributes specifying whether they are bold or italic.
3. Evaluation
In this section we present the evaluation of our reference extraction algorithm. We first evaluate the retrieval of citation strings resulting from reference segmentation, then we continue with the results of the classification of reference tokens.
3.1 Data Set
We use a dataset of documents from PubMed, a free database created by the US National Library of Medicine holding full-text articles from the biomedical domain, together with a standard XML markup that rigorously annotates the complete content of the published document, including reference meta-data. Note that this dataset differs substantially from the dataset on which the original ParsCit system was trained. While it consists of mostly journal articles from the biomedical domain, ParsCit focused on computer science papers, mostly from conferences [4].
The PubMed dataset consists of 10,000 documents for training and 9,581 documents for testing. Of these, 4,602 training documents and 4,469 test documents contained at least one reference as ground truth. For about 98.5% of the documents we were able to extract at least one reference via our reference line extraction. Most of the remaining documents are articles from journals whose layout does not include a reference header (e.g., BMJ). The following evaluation includes only those documents which contain at least one reference in the ground truth and from which at least one reference can be extracted.
3.2 Reference Segmentation
In order to evaluate the reference segmentation, we have to map each extracted reference string to the corresponding ground truth reference. This is done in the following manner. First, the author, title, and year information of each ground truth reference from PubMed is tokenized. Then, for each extracted reference string we count how many tokens match, and choose the one with the highest overlap. Additionally, for a successful mapping, the overlap needs to be at least half the number of tokens of the PubMed reference.
The results of the reference segmentation are shown in Table 1 for the training set, and Table 2 for the test set. Precision, recall, and F1 values are determined based on the number of matched, spurious, and missing reference strings resulting from the reference mapping with the ground truth. Three scenarios are compared including increasing levels of layout information: (i) citations extracted by the out-of-the-box ParsCit system applied to the raw text output of PDFBox, (ii) reference segmentation with PDF preprocessing (i.e., detection of columns/blocks, reading order, and decorations), and (iii) reference segmentation with PDF pre-processing and additional layout information (i.e., line gaps and indentations). It can be seen that the PDF preprocessing improves the F1 of retrieved reference strings from about 88% to 90%, and the inclusion of additional layout information further improves the F1 by another 3%. This means that reference segmentation can be considerably improved by using various types of layout information.
Table 1: Results of the reference segmentation on the PubMed training dataset. |
| Configuration |
Precision |
Recall |
F1 |
| oob. ParsCit on PDFBox |
90.7% |
85.9% |
88.2% |
| w. PDF prepr. |
91.0% |
89.5% |
90.3% |
| w. PDF prepr. & layout |
93.1% |
94.6% |
93.8% |
Table 2: Results of the reference segmentation on the PubMed test dataset. |
| Configuration |
Precision |
Recall |
F1 |
| oob. ParsCit on PDFBox |
90.1% |
85.1% |
87.5% |
| w. PDF prepr. |
91.1% |
90.0% |
90.5% |
| w. PDF prepr. & layout |
92.6% |
94.9% |
93.7% |
3.3 Reference Meta-data Classification
The classification of individual reference meta-data fields, such as title, journal or source, issue, volume, etc., is performed on the token level. Thus, for the evaluation, each token has to be assigned a corresponding label from the ground truth. We have already aligned the extracted reference strings with the corresponding ground truth references; tokens therefore need to be matched only within such a reference pair. This is done by simply searching each extracted token in the ground truth, and assigning the corresponding ground truth label. If a token is not found in the ground truth, or found multiple times, the label is determined by the neighborhood of labels. For example, if a subsequence of labels is title, title, X, title, and token X is not found in the ground truth, it is assigned the label title. Similarly, the token X in authorGivenName, X, authorSurname is assigned the label authorOther.
We start with the evaluation of the out-of-the-box ParsCit system applied to the raw text obtained by PDFBox. The result of the ParsCit executable is an XML file with a list of citations with the fields author, booktitle, date, editor, institution, journal, location, note, pages, publisher, tech, title, volume, issue, and number. These fields are aligned to the PubMed fields in a straightforward manner (journal is equal to source, and author names are combined into a single author label). The resulting evaluation is shown in Table 3. It can be seen that the retrieval of author, title, and date tokens works reasonably well, however, the performance for most other token types could be improved. The main reason for that is that the out-of-the-box ParsCit system was trained on a set of documents with rather different characteristics.
Table 3: Results of the reference token classification of the out-of-the-box ParsCit system applied to the raw text obtained by PDFBox on the PubMed test dataset. |
| Meta-data |
Precision |
Recall |
F1 |
| date |
0.98 |
0.92 |
0.95 |
| author |
0.92 |
0.90 |
0.91 |
| title |
0.79 |
0.95 |
0.86 |
| volume |
0.91 |
0.31 |
0.46 |
| pages |
0.54 |
0.51 |
0.53 |
| source |
0.93 |
0.48 |
0.64 |
| editor |
0.22 |
0.21 |
0.22 |
| issue |
0.80 |
0.26 |
0.39 |
| publisher |
0.85 |
0.27 |
0.41 |
| location |
0.24 |
0.64 |
0.35 |
We continue with the evaluation of the reference token classification using the original ParsCit feature set. The performance values for individual token types are shown in Table 4, where the sequence classification method used is Maximum Entropy with Beam Search. Most token types are recognized with an F1 of at least 0.8, and only for labels which occur more rarely, such as edition or conference, is the performance is worse. The substantial improvement compared to the performance of the out-of-the-box ParsCit system shown in Table 3 is due to the fact that the training set consisted of a much larger number of documents covering a wide range of layouts of articles from the biomedical domain, whereas the out-of-the-box ParsCit system was trained on a comparably limited number of computer science papers. This demonstrates the importance of the adaptation of the classification model to the right document statistics.
Table 4: Results of the reference token classification using the original ParsCit feature set trained with Maximum Entropy & Beam Search on the PubMed dataset. |
| Meta-data |
Precision |
Recall |
F1 |
| date |
0.97 |
0.98 |
0.98 |
| authorGivenName |
0.95 |
0.99 |
0.97 |
| authorSurname |
0.95 |
0.99 |
0.97 |
| title |
0.95 |
0.98 |
0.97 |
| volume |
0.97 |
0.94 |
0.96 |
| pages |
0.96 |
0.95 |
0.96 |
| source |
0.96 |
0.92 |
0.94 |
| note |
0.79 |
0.93 |
0.86 |
| editor |
0.92 |
0.79 |
0.85 |
| issue |
0.93 |
0.78 |
0.85 |
| publisher |
0.89 |
0.63 |
0.74 |
| location |
0.75 |
0.46 |
0.57 |
| edition |
0.69 |
0.11 |
0.19 |
| conference |
0.92 |
0.07 |
0.13 |
Table 5 shows the same performance values if we additionally include font information, i.e., whether a token has a different font than the tokens inside a sliding window neighborhood from -2 to +2 tokens. It can be seen that the recognition of some token types can be slightly improved. For example, volume or issue numbers are sometimes printed in bold or italic font, and the classifiers are able to make use of this additional information.
Table 5: Results of the reference token classification including additional font information trained with Maximum Entropy & Beam Search on the PubMed dataset. |
| Meta-data |
Precision |
Recall |
F1 |
| date |
0.99 |
0.99 |
0.99 |
| authorGivenName |
0.94 |
0.99 |
0.96 |
| authorSurname |
0.95 |
0.99 |
0.97 |
| title |
0.96 |
1.00 |
0.98 |
| volume |
0.98 |
0.98 |
0.98 |
| pages |
0.95 |
0.96 |
0.95 |
| source |
0.96 |
0.93 |
0.95 |
| note |
0.91 |
0.83 |
0.87 |
| editor |
0.90 |
0.84 |
0.87 |
| issue |
0.93 |
0.81 |
0.86 |
| publisher |
0.88 |
0.68 |
0.76 |
| location |
0.80 |
0.58 |
0.67 |
| edition |
0.59 |
0.10 |
0.17 |
| conference |
0.81 |
0.07 |
0.13 |
Figure 1 summarizes the F1 values obtained on individual token labels for all three scenarios: out-of-the-box ParsCit, ParsCit retrained on a representative training set using the original features, and ParsCit retrained on an enhanced feature set including additional font information.
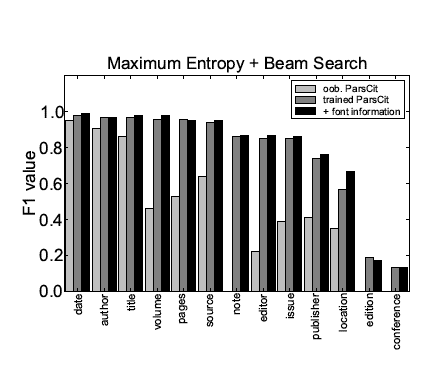 Figure 1: Comparison of the performance of the reference token classification
Figure 1: Comparison of the performance of the reference token classification
using Maximum Entropy & Beam Search for different meta-data fields and different scenarios.
Tables 6 and 7 show the performance values obtained with CRFs for both scenarios, and Figure 2 shows a direct comparison. The overall performance is similar as in the case of Maximum Entropy with Beam Search. Again, the retrieval of some token types can be improved by incorporating additional font information.
Table 6: Results of the reference token classification using the original ParsCit feature set trained with CRF on the PubMed dataset. |
| Meta-data |
Precision |
Recall |
F1 |
| date |
0.98 |
0.99 |
0.98 |
| authorGivenName |
0.95 |
0.98 |
0.97 |
| authorSurname |
0.95 |
0.98 |
0.97 |
| title |
0.96 |
0.97 |
0.97 |
| volume |
0.97 |
0.93 |
0.95 |
| pages |
0.95 |
0.96 |
0.95 |
| source |
0.96 |
0.94 |
0.95 |
| note |
0.80 |
0.89 |
0.84 |
| editor |
0.82 |
0.83 |
0.83 |
| issue |
0.94 |
0.70 |
0.80 |
| publisher |
0.83 |
0.83 |
0.83 |
| location |
0.62 |
0.47 |
0.53 |
| edition |
0.69 |
0.11 |
0.19 |
| conference |
0.40 |
0.14 |
0.21 |
Table 7: Results of the reference token classification including additional font information trained with CRF on the PubMed dataset. |
| Meta-data |
Precision |
Recall |
F1 |
| date |
0.99 |
0.99 |
0.99 |
| authorGivenName |
0.95 |
0.98 |
0.97 |
| authorSurname |
0.95 |
0.99 |
0.97 |
| title |
0.98 |
0.99 |
0.98 |
| volume |
0.99 |
0.98 |
0.98 |
| pages |
0.95 |
0.97 |
0.96 |
| source |
0.96 |
0.96 |
0.96 |
| note |
0.88 |
0.88 |
0.88 |
| editor |
0.85 |
0.87 |
0.86 |
| issue |
0.93 |
0.82 |
0.87 |
| publisher |
0.84 |
0.90 |
0.87 |
| location |
0.76 |
0.59 |
0.67 |
| edition |
0.26 |
0.10 |
0.15 |
| conference |
0.57 |
0.11 |
0.18 |
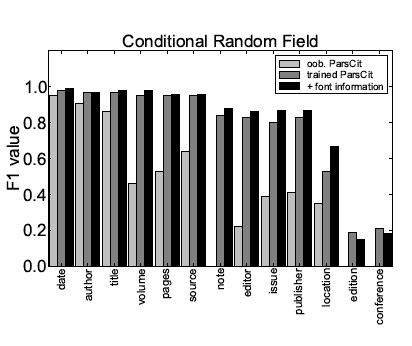 Figure 2: Comparison of the performance of the reference token classification
Figure 2: Comparison of the performance of the reference token classification
using CRF for different meta-data fields and different scenarios.
These findings are generally in line with the results in [12]. One exception is the location field, for which [12] reports an F1 of 0.87, whereas in Tables 4-7 values from 0.57-0-67 are achieved. One reason for this is that location might be undersampled in the biomedical domain, where references mostly consist of journal publications. The dataset in [12] is from the computer science domain, where the location field occurs more frequently in citations of conference proceedings, books, or book chapters.
4. Discussion
Our results show that the consideration of additional layout and formatting information available from PDF files improves the performance of the individual stages of a reference extraction system. Previous studies have used similar layout and formatting information in order to extract features for supervised extraction of meta-data other than references [6, 8, 12]. In [8] features are extracted from text blocks providing information about the position of a block within a page and of a token within a block, as well as their alignment and formatting of fonts. Similar to ParsCit, [12] uses raw text as input; it is shown there that the inclusion of information about line breaks and about the position of a word within a line considerably improves the meta-data extraction performance. The layout-based approach described in [6] also operates on raw text, but uses information about the font and the position of tokens provided by the PDF.
In all of these approaches meta-data are extracted in a single classification step using quite generic features. In contrast, our algorithm consists of several sequential stages where different layout and formatting features are employed, which are designed specifically for the extraction of references. The reference line extraction stage exploits information about columns, text blocks, and vertical gaps between lines; the reference segmentation stage uses information about line indentations; the reference token classification utilizes font information. Information about line indentations and vertical gaps between lines is usually not available in systems which take the raw text as input, such as the original ParsCit or [12]. The approach in [6] extracts the raw text from a PDF, but does not attempt to recognize columns or the reading order of the text. In contrast to [8], where text blocks were extracted and directly classified, we used this information in the preceding reference line extraction and reference segmentation stages.
Our system is based on the open source Java library PDFBox, which yields a text stream of individual characters and their positions on the page. One problem with this and other tools, however, is that the information provided about individual characters in the PDF is inherently noisy, for example, height and width information might be wrong, or information about the font of some characters might be missing. This implicit noise affects every stage of our system, and we believe that its performance could be considerably improved if this low-level information would be more reliable.
Other common sources of error include the following items:
- Authors may be composed of tokens none of which are names, e.g., "American Medical Association Council on Medical Education". This affects the recognition of authors because ParsCit uses a list of common names.
- The same journal name might be written in different ways, e.g., "NZ Med J" vs "N Z Med J".
- Some parts of the volume field is missing, e.g., "58:" vs "58(1):".
- Often there is a lot of added noise after a reference string, e.g., "(article in German)". This complicates the reference mapping process.
- Sometimes references span multiple pages, but there were errors in the PDF extraction, e.g., decorations were not properly detected or there are other rendering issues.
- The optimal handling of field separators is still an open question, and better heuristics to split the reference string into its tokens could further improve the performance of the token classification. For example, authors are often separated from the title with a colon.
5. Conclusions
In this paper we have quantified the impact of adding layout and formatting information available in PDF files on the performance of ParsCit, a state-of-the-art system for extracting reference meta-data from scientific articles. This additional information included the boundaries of columns, the reading order of text blocks on a page, the vertical gaps between lines and their indentation, as well as the font information of individual tokens within the reference string. The inclusion of these points into the reference extraction algorithm resulted in both a better reference segmentation and token classification. By retraining ParsCit on the PubMed dataset, we also demonstrated the effect of using a larger, more diverse, and more representative training set.
In future work we plan to take into account the layouts of individual journals. One way would be to train different classifiers for different journals, for which we could use the journal information that we extract using our previous work on meta-data extraction [8]. Another approach would be to cluster reference styles in an unsupervised manner.
Acknowledgements
The presented work was in part developed within the CODE project funded by the EU FP7 (grant no. 296150) and the TEAM IAPP project (grant no. 251514) within the FP7 People Programme. The Know-Center is funded within the Austrian COMET Program — Competence Centers for Excellent Technologies — under the auspices of the Austrian Federal Ministry of Transport, Innovation and Technology, the Austrian Federal Ministry of Economy, Family and Youth and by the State of Styria. COMET is managed by the Austrian Research Promotion Agency FFG.
References
[1] M. Aiello, C. Monz, L. Todoran, and M. Worring. Document understanding for a broad class of documents. International Journal on Document Analysis and Recognition, 5(1):1-16, 2002.
[2] A. L. Berger, V. J. D. Pietra, and S. A. D. Pietra. A Maximum Entropy Approach to Natural Language Processing. Computational Linguistics, 22(1):39-71, 1996.
[3] E. Cortez, A. S. da Silva, M. A. Goncalves, F. Mesquita, and E. S. de Moura. FLUX-CIM: flexible unsupervised extraction of citation metadata. In JCDL '07: Proceedings of the 2007 Conference on Digital Libraries, pages 215-224, 2007. http://doi.org/10.1145/1255175.1255219
[4] I. G. Councill, C. L. Giles, and M.-y. Kan. ParsCit: An open-source CRF Reference String Parsing Package. In N. Calzolari, K. Choukri, B. Maegaard, J. Mariani, J. Odjik, S. Piperidis, and D. Tapias, editors, Proceedings of LREC, volume 2008, pages 661-667. Citeseer, European Language Resources Association (ELRA), 2008.
[5] C. Giles, K. Bollacker, and S. Lawrence. CiteSeer: An automatic citation indexing system. In Proceedings of the third ACM conference on Digital Libraries, pages 89-98, 1998. http://doi.org/10.1145/276675.276685
[6] M. Granitzer, M. Hristakeva, R. Knight, K. Jack, and R. Kern. A Comparison of Layout based Bibliographic Metadata Extraction Techniques. In WIMS'12 — International Conference on Web Intelligence, Mining and Semantics, pages 19:1-19:8. ACM New York, NY, USA, 2012. http://doi.org/10.1145/2254129.2254154
[7] I.-a. Huang, J. Ho, H. Kao, and W. Lin. Extracting citation metadata from online publication lists using BLAST. In Proc. of the 8th Pacific-Asia Conference on Knowledge Discovery and Data Mining, 2004. http://doi.org/10.1007/978-3-540-24775-3_64
[8] R. Kern, K. Jack, M. Hristakeva, and M. Granitzer. TeamBeam — Meta-Data Extraction from Scientific Literature. D-Lib Magazine, Vol. 18, No. 7/8, 2012. http://doi.org/10.1045/july2012-kern
[9] J. Lafferty, A. Mccallum, F. Pereira, and I. Science. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In Proceedings of the International Conference on Machine Learning (ICML-2001), 2001.
[10] X. Lin. Header and Footer Extraction by Page Association. Proceedings of SPIE, 5010:164-171, 2002. http://doi.org/10.1117/12.472833
[11] M.-T. Luong, T. D. Nguyen, and M.-Y. Kan. Logical structure recovery in scholarly articles with rich document features. International Journal of Digital Library Systems, 1(4):1-23, Jan. 2011. http://doi.org/10.4018/jdls.2010100101
[12] F. Peng and A. McCallum. Accurate Information Extraction from Research Papers using Conditional Random Fields. In HLTNAACL04, volume 2004, pages 329-336, 2004.
[13] A. Ratnaparkhi. Maximum Entropy Models for Natural Langual Ambiguity Resolution. PhD thesis, 1998.
[14] K. Seymore, A. McCallum, and R. Rosenfeld. Learning Hidden Markov Model Structure for Information Extraction. In AAAI 99 Workshop on Machine Learning for Information Extraction, pages 37-42, 1999.
About the Authors
 |
Roman Kern is senior researcher at the Graz University of Technology, where he works on information retrieval, social web and natural language processing. In addition he has a strong background in machine learning, which he acquired during his work at the Know-Center. Previous to working in research he gained experience in industry projects at Hyperwave and DaimlerChrysler. There he worked as project manager, software architect and software engineer for a couple of years. He obtained his Master's degree (DI) in Telematics at the Technical University of Graz. He published over 20 peer-reviewed publications and achieved top rank results in international scientific challenges like CLEF, ACL SemEval. Most of his publication work can be accessed online: http://www.mendeley.com/profiles/roman-kern/.
|
 |
Stefan Kampfl is a postdoctoral researcher at the Know-Center, with expertise in the fields of machine learning and data analysis. He received both his DI degree (in Telematics; equivalent to an MSc in information and communication technology) and his PhD (in computational neuroscience) at Graz University of Technology. In his PhD thesis he investigated unsupervised machine learning algorithms and their role in information processing in the brain, and analyzed biological data using methods from machine learning and information theory. While writing his thesis, he contributed to a number of high-impact conference and journal publications.
|
|