 |
D-Lib Magazine
July/August 2012
Volume 18, Number 7/8
Table of Contents
TeamBeam — Meta-Data Extraction from Scientific Literature
Roman Kern
Graz University of Technology, Graz, Austria
rkern@tugraz.at
Kris Jack
Mendeley Ltd.
kris.jack@mendeley.com
Maya Hristakeva
Mendeley Ltd.
mhristakeva@gmail.com
Michael Granitzer
University of Passau, Passau, Germany
michael.granitzer@uni-passau.de
doi:10.1045/july2012-kern
Printer-friendly Version
Abstract
An important aspect of the work of researchers as well as librarians is to manage collections of scientific literature. Social research networks, such as Mendeley and CiteULike, provide services that support this task. Meta-data plays an important role in providing services to retrieve and organise the articles. In such settings, meta-data is rarely explicitly provided, leading to the need for automatically extracting this valuable information. The TeamBeam algorithm analyses a scientific article and extracts structured meta-data, such as the title, journal name and abstract, as well as information about the article's authors (e.g. names, e-mail addresses, affiliations). The input of the algorithm is a set of blocks generated from the article text. A classification algorithm, which takes the sequence of the input into account, is then applied in two consecutive phases. In the evaluation of the algorithm, its performance is compared against two heuristics and three existing meta-data extraction systems. Three different data sets with varying characteristics are used to assess the quality of the extraction results. TeamBeam performs well under testing and compares favourably with existing approaches.
Keywords: Meta-data Extraction, Supervised Machine Learning, Content Analysis and Indexing, Natural Language Processing, Text Analysis
1 Introduction
In recent years social research networks have gained a lot of momentum. Researchers are able to manage their collection of scientific articles and exchange and discuss papers with colleagues. Examples of such research networks are Mendeley and CiteULike. The quality of services provided by such systems depends on the information that can be extracted from articles. In the traditional ecosystem, publishers could afford to manually extract the relevant meta-data or impose this task on the authors of the articles. In the world of collaborative research networks, however, these tasks are typically crowdsourced. This process can be boot-strapped by extracting the meta-data via tools, which are able to automatically retrieve relevant information. Furthermore, a crowdsourcing approach will work less well when applied to the long tail of articles that have few readers.
The TeamBeam algorithm has been developed to provide a flexible tool to extract a wide array of meta-data from scientific articles. At its core, TeamBeam is a supervised machine learning algorithm, where labelled training examples are used to learn a classification scheme for the individual text elements of an article. Performance can be improved by integrating sequence information into the classification process.
This approach has already been followed in the past, as some classification algorithms already integrate sequence into their model, making them viable candidates for this task. Typical examples of this family of classification algorithms are Hidden Markov Models (HMMs) [9] and Conditional Random Fields (CRFs) [9]. CRFs tend to form the base of many existing meta-data extraction approaches [2][3][6]. Classification algorithms, which do not have a native support for sequences, can be enhanced to improve the classification results by additional logic. For example Support Vector Machines (SVMs) have been utilised for the task of meta-data extraction [4][5]. This approach is followed by meta-data extraction tools employed by Mendeley Desktop and CiteSeer platform.
The TeamBeam algorithm uses a Maximum Entropy [1] classifier, which is enhanced by the Beam Search [8] for the sequence classification task. This combination has already been applied in the area of Natural Language Processing to label sequences of words. In terms of feature types, the TeamBeam algorithm follows existing approaches and integrates layout and formatting information as well as employing common name lists. It goes beyond existing approaches by creating language models [7] during the training phase.
2 Algorithm
The TeamBeam algorithm takes a scientific article as input and extracts a number of meta-data fields (e.g. title, author names). A demonstration of the algorithm can be accessed online (Figure 1). The source of the algorithm is available under an open-source license, which can an be accessed via the TeamBeam web-page.
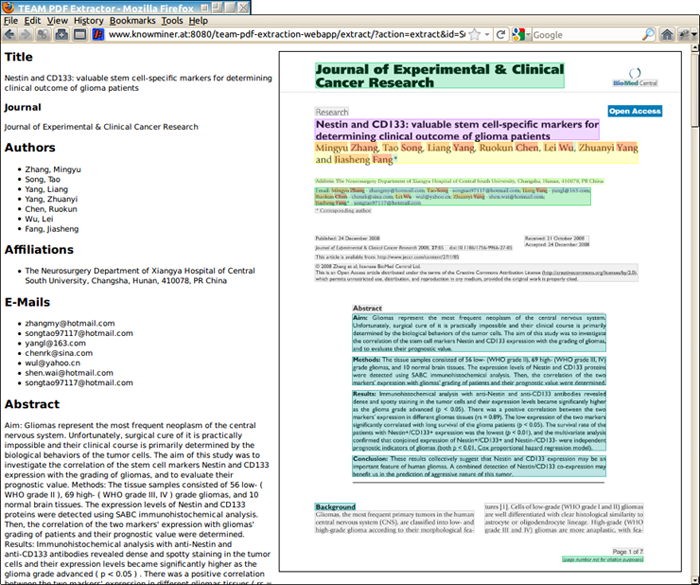 Figure 1: Example of a scientific article together with the output of the TeamBeam algorithm. Meta-data has been extracted and also annotated in the preview image of the article.
Figure 1: Example of a scientific article together with the output of the TeamBeam algorithm. Meta-data has been extracted and also annotated in the preview image of the article.
Input: The starting point for meta-data extraction is a set of text blocks, which are provided by an open-source tool, which can also be accessed via the TeamBeam web-page, that is built upon the output of the PDFBox library. These blocks are generated from parsing scientific articles and organising the text into words, lines and then text blocks. To identify these text blocks, layout and formatting information is exploited, where a text block is a list of vertically adjacent lines which share the same font size. For illustration purposes the input for the meta-data extraction can be expressed as XML:
<document pages="8">
<page width="21.0" height="29.7">
<block x="2.1" y="10.3" width="15.3" height="2.2" font-size="12">
<line>
<word>Nestin</word>
<word>and</word>
<word>CD133</word>
</line>
</block>
</page>
</document>
Processing Pipeline: The extraction process consists of two consecutive classification phases, text block classification and token classification. In the first phase the input text blocks are classified and a sub-set of the meta-data are derived from this result. The text blocks related to the author meta-data are then fed into another classification phase. In the second phase the individual words within the text blocks are classified.
Meta-Data Types: The goal of the TeamBeam algorithm is to extract a rich set of meta-data from scientific articles: i) The title of the scientific article; ii) The optional sub-title, which is only present for a fraction of all available articles; iii) The name of the journal, conference or venue; iv) The abstract of the article, which might span a number of paragraphs; v) The names of the authors; vi) The e-mail addresses of the authors; vii) The affiliation (i.e. university, institute, company, etc.) of the authors.
Classification Algorithm: A supervised machine learning algorithm lies at the core of the meta-data extraction process. The open-source library OpenNLP provides a set of classification algorithms tailored towards the classification of sequences. Its main algorithm is based on the Maximum Entropy classifier [1], which by itself does not take sequence into account. In order to integrate the sequence information, a Beam Search approach [8] is followed. Beam Search takes the classification decision of preceding instances into account to improve overall performance and to rule out any unlikely label sequences.
Feature Types: Classification algorithms are capable of dealing with a diverse set of feature types. The TeamBeam algorithm is restricted to binary features. Therefore, all continuous or categorical information needs to be mapped to features with binary values. The features used for classification are derived from the layout, the formatting, the words within and around a text block, and common name lists. A language model is created at the beginning of the training phase to improve the text block classification.
There are two common name lists which are used to create features for the classification: a list of common first names and a list of surnames. The first list contains 7,133 entries, which represent common first names taken from the GATE project. The list of surnames contains 88,799 names which represent the most common names from the US Census. Both lists are biased towards US names and therefore cannot be seen as exhaustive.
2.1 Text Block Classification
The first phase of meta-data extraction is the classification of the text blocks.
Its task is to assign each text block on a page to one of these labels: Title Block; Sub-Title Block; Journal Block; Abstract Block; Author Block; E-Mail Block; Affiliation Block; Author-Mixed Block; and Other Block.
Depending on the layout of the input article, the author related meta-data may either be found in separate text blocks, or a single block may contain more than one author meta-data type.
In the latter case, the block should be labelled with Author-Mixed Block, for example if the e-mail address and the affiliation occur in the same text block.
The Other Block class is assigned to all text blocks that contain no meta-data.
The text block label from the training set is also used to build a language model. For each block type, all of the words contained within are collected and their frequency is counted.
This is done for all articles from the training set. Thus each text block type has its own language model, where the frequency of words reflect how often this word has been used within such a text block.
- Language Model Features
- When creating the feature set for each text block, all words within the block are iterated over. For each word, the text block type with the highest likelihood according to the language models is counted. The text block type with the highest count is then assumed to be the most probable source for the words contained within a single text block. This is the base for the first set of features.
- Layout Features
- The set of binary features which encode the position of a single block within a page are:
isFirstBlock; isLastBlock; isLeftHalf; isRightHalf; isTopHalf; isBottomHalf; isRight; isLeft; isTop; isBottom; and isCenter.
In addition there are two sets of fuzzy isRight, isLeft, isTop and isBottom features, which are generated for two different relative thresholds from their respective page border (0.1 and 0.3).
- Formatting Features
- The formatting features encode font and text flow in the text:
isBigFont; isBiggerFont; isSmallFont; isSmallerFont; isLeftAligned; and isRightAligned.
The reference for the font size is computed by the mean font size plus/minus the standard deviation, which is multiplied by 1 for Big/Small and 1.5 for Bigger/Smaller. A set of features is created for the number of characters per line and the number of lines within each block, once for the count as is and once for the floored square root of the count.
- Dictionary Features
- If a word within a text block is found in one of the two common name lists, a binary feature is added to the feature set: containsGivenName; and containsSurname.
- Heuristic Features
- There are a number of simple heuristic features: containsEMail; containsAtChar; containsDigits; and containsPunctuation.
To detect e-mail addresses, a regular expression is applied on the text.
- Term Features
- All words with a block are also added to the feature space, after being converted to lower case. The first and last word of the block directly to the right, top, left and bottom of the current block are added.
OpenNLP provides the functionality to add another set of features specifically for the Beam Search algorithm, in which the labelling output of the two preceding text blocks, as well as the labelling of the block above, right and left of the current block, is added. As the blocks are vertically sorted, either the right or the left block information might not be available.
2.2 Token Classification
The output of the first classification phase is then used as starting points for the second phase. All text blocks labelled with one of the author related types are further processed.
The target of the second classification run is the individual words within the text blocks. These are assigned to one of these classes: Given Name; Middle Name; Surname; Index; Separator; E-Mail; Affiliation-Start; Affiliation; and Other.
The affiliation tokens are split into two classes, one for the initial word of an affiliation. This is motivated by the need to separate two different affiliations, which are written in a sequence. Furthermore, one can assume that the initial word of an affiliation often will be from a small set of common words, for example "University" or "Institute". The index class is used for special characters that link an author name to a corresponding e-mail address and affiliation. Common index characters are an asterisk and numbers, which are often formatted as superscript. The separator class is used to separate multiple index characters, usually a comma.
The features for the token classification are less rich than the text block features.
- Language Model Features
- The relative frequency of words is reflected in three features: isCommonWord ( > 0.1); isInfrequentWord ( < 0.01); and isRareWord ( < 0.001).
- Layout Features
- Three features indicate the token's position: isFirstInLine; isFirst; and isLast
- Formatting Features
- Again, the average font size in combination with the standard deviation is used for: isBigger; and isSmaller.
In addition, the number of characters within the token is added as a feature.
- Dictionary Features
- These features are set if the token occurs in the list of given names, or surname lists: containsGivenName; and containsSurname
- Heuristic Features
- The heuristic features from the text block classification are re-used for the token classification.
In addition, a feature is generated for initials (upper-case character followed by a dot).
- Term Features
- The token itself is converted into a feature, after being normalised to lower-case.
As additional input for the Beam Search, the labelling decision of the preceding three tokens is added to the set of features.
2.3 Meta-data Extraction
Some of the meta-data types can be taken directly from the output of the text block classification.
Here, all blocks of a specific class are concatenated to form a single meta-data value.
These meta-data types are: Title; Sub-Title; Journal; and Abstract.
To extract the other meta-data types, the output of the token classification is processed.
The meta-data extracted at this phase are: Author (Given Name, Middle Name and Surname); E-Mail; and Affiliation.
3 Data Sets
In the evaluation section TeamBeam is applied to three different data sets. The three differ in the selection of scientific articles and journals. They also vary in layout and formatting, as well as in their topics. Two of the three data sets are generated from publicly available sources, so the results can be independently verified.
- E-Prints
- This data set was generated by crawling the RDF repository of the RKB-Explorer project.
All articles were included from journals and conferences with more than 10 assigned papers.
The final data set contains 2,452 PDFs together with the meta-data collected from the RDF.
It covers a wide range of layout styles from a diverse set of domains.
- Mendeley
- The next data set was created from the article catalogue of the Mendeley research network.
It contains 20,672 articles sampled from a range of different domains.
The meta-data for these papers is manually curated and crowdsourced by the Mendeley users.
As it may also contain articles not published under an open access license, this data set cannot be made available for download.
- PubMed
- The final data set was created from the public articles available from PubMed.
It matches the Mendeley data set in terms of size and contains 19,581 entries.
This data set provides a rich set of meta-data.
In contrast to the other two data sets, it also contains information about the e-mail addresses and affiliation of the authors.
The article collection also contains a number of older publications, which have been scanned in and processed with OCR software.
Furthermore, the data set represents a range of different article types, for example book reviews and product presentations.
All three data sets contain articles with different layout and formatting. The meta-data gathered for the E-Prints data set contains the highest level of noise, where author names and even titles are sometimes abbreviated.
4 Evaluation
In this section, the performance of the presented approach is evaluated against the three data-sets. Its performance is compared to a simple baseline and alternative, existing approaches. For the evaluation, the TeamBeam algorithm is only applied on the first page of an article. All other pages are ignored. As the data sets contain too few examples for the sub-title meta-data, this type is not included in the presented evaluation.
4.1 Baselines
To put the measured performance of the meta-data extraction approaches into perspective, the performance of two simple heuristics is presented first.
- Title Heuristic
-
The first heuristic tries to identify the title meta-data. The font size of all text blocks on the first page are inspected. The blocks are then sorted by decreasing font size. For some journals the name of the journal is larger than the title, so another constraint is introduced. Typically the journal names are found at the very top of the page. Therefore, all blocks are discarded which overlap with the top 20% of the page.
- E-Mail Heuristic
-
The second heuristic tries to identify the e-mail addresses of the authors. A regular expression was constructed, which matches valid e-mail addresses. In contrast to the first heuristic, the e-mail identification operates on plain text instead of blocks of text.
The two heuristics were applied to a set of randomly selected articles from the PubMed data set. The test data set for applying the heuristics is made up of 2,012 articles. The results of applying these simple heuristics are already quite good but still leave room for improvement (Table 1).
| Heuristic |
Precision |
Recall |
|
| Title |
0.62 |
0.62 |
|
| E-Mail |
0.84 |
0.64 |
|
Table 1: Performance of two simple heuristics on a subset of the PubMed data set.
4.2 Comparison with Existing Algorithms
Besides the simple heuristics, previous work with alternative approaches is presented for comparison. As the performance of information extraction algorithms is dependent on the data sets, we have limited the comparison to the evaluations done on the data sources E-Print and Mendeley. Still, there are three different approaches that differ in terms of features, machine learning algorithms and text extraction pipelines.
- ParsCit
- The ParsCit approach [2] employs a classification scheme based on Conditional Random Fields.
Its features do not exploit the layout information, but it uses look-up tables and dictionaries.
Besides the supervised machine learning algorithm, ParsCit does include a number of heuristics to improve its performance.
The classification model has not been retrained to match the test data set presented here.
- Layout-CRFs
- The next approach is similar to ParsCit as it also uses Conditional Random Fields as the classification algorithm but, in contrast to ParsCit, it does not use dictionaries nor look-up tables, instead integrating the layout information [3].
Here, performance depends on the classification algorithm, as no further heuristics or manually created constraints are applied.
- Mendeley Desktop
- Support Vector Machines are central to the Mendeley meta-data extraction process.
The classification has been enhanced by including formatting and contextual features [4].
Again, heuristics have been developed to enhance the performance of classification results.
The approaches compared differ in terms of the feature types that they use and classification algorithms employed (Table 2). The TeamBeam algorithm exploits layout information and contains dictionaries for names. It also includes a regular expression heuristic for e-mail addresses, which cannot be compared in level of sophistication with the ParsCit and Mendeley Desktop heuristics.
| Approach |
H |
D |
L |
M |
Algorithm |
|
| ParsCit |
• |
• |
|
|
CRF |
|
| Layout-CRF's |
|
|
• |
|
CRF |
|
| Mendeley Desktop |
• |
• |
• |
|
SVM |
|
| TeamBeam |
 |
• |
• |
• |
MaxEnt |
|
Table 2: Comparison of features types between the four different approaches. Feature types are heuristics (H), dictionaries (D), layout information (L) and language model (M).
The numbers reported for the three existing approaches can be seen as a guide and cannot directly be compared to the performance of the TeamBeam algorithm. The evaluations use different splits between training and test data and differ in the fuzziness of string comparisons. For example, the numbers reported for the reference algorithms are computed using a more fuzzy string comparison metric, allowing a maximum Levenshtein distance of up to 0.7, whereas in the evaluation of TeamBeam the maximum edit distance is less lenient, max(1, ⎣#characters / 10 ⎦).
Most importantly, however, the performance of the meta-data extraction process heavily depends on the preceding text processing. Unfortunately, for many PDFs the results of the text extraction and the extraction of layout information is not identical between different implementations. Mendeley Desktop employs PDFNet to convert PDFs into text, while all other approaches are based on the output of the open-source library PDFBox. Errors introduced during the text extraction process are propagated and can impact the classification results.
When comparing the three approaches with each other, one can see that the layout information is helpful for some meta-data, but does not appear to be relevant for other meta-data. The Layout-CRF's algorithm outperforms the ParsCit approach for titles, while the dictionary based approach of ParsCit performs better for author names on the E-Prints data set (Table 3).
| Meta-Data |
Approach |
Precision |
Recall |
|
| Title |
ParsCit |
0.65 |
0.61 |
|
| Layout-CRF's |
0.75 |
0.83 |
|
| Mendeley Desktop |
0.61 |
0.49 |
|
| Author |
ParsCit |
0.59 |
0.39 |
|
| Layout-CRF's |
0.50 |
0.29 |
|
| Mendeley Desktop |
0.53 |
0.29 |
|
Table 3: Overview of the performance of various meta-data extraction algorithms on the E-Prints data set.
The Mendeley data set includes different journals so there is a variety of layouts and formatting characteristics across the articles. The performance numbers therefore vary in comparison to the E-Prints data set. The Mendeley Desktop algorithm demonstrates the best performance, indicating that the algorithm might have been tuned for this data set (Table 4).
| Meta-Data |
Approach |
Precision |
Recall |
|
| Title |
ParsCit |
0.75 |
0.69 |
|
| Layout-CRF's |
0.73 |
0.73 |
|
| Mendeley Desktop |
0.94 |
0.91 |
|
| Author |
ParsCit |
0.77 |
0.50 |
|
| Layout-CRF's |
0.50 |
0.21 |
|
| Mendeley Desktop |
0.81 |
0.62 |
|
Table 4: Performance of the existing algorithms on the Mendeley data set. The Mendeley Desktop algorithm provides the best performance.
4.3 Text Block Classification
The evaluation of the text block classification is conducted on all three data sets, where 1,000 articles are randomly selected for training and another 1,000 for testing (Table 5). Only the PubMed data set contains information about the author e-mail and affiliation. One can see that the classification itself does provide satisfactory results, with most of the precision and recall figures being close to 0.9. The only exception is the abstract block type, where the recall falls behind the results of the other text block types. Closer inspection indicates that in cases where the abstract is spread over multiple text blocks only a subset of them are successfully found in the test set.
| |
E-Prints |
Mendeley |
PubMed |
| Text-Block Type |
Precision |
Recall |
Precision |
Recall |
Precision |
Recall |
| Title Block |
0.94 |
0.86 |
0.98 |
0.97 |
0.95 |
0.92 |
| Journal Block |
0.90 |
0.87 |
0.97 |
0.94 |
0.82 |
0.82 |
| Abstract Block |
0.89 |
0.46 |
0.89 |
0.62 |
0.94 |
0.85 |
| Author Block |
0.90 |
0.76 |
0.95 |
0.89 |
0.94 |
0.87 |
| E-Mail Block |
|
|
|
|
0.95 |
0.91 |
| Affiliation Block |
|
|
|
|
0.88 |
0.87 |
Table 5: Evaluation results for the three data sets for the text block classification stage. The numbers reported here do not include the errors introduced in the text processing stage.
4.3.1 Comparison of Algorithms
The open-source machine learning framework Weka provides a rich set of classification algorithms. This allows us to compare different approaches with each other (Table 6). If not otherwise specified, the Weka default values for the algorithm parameters were used. In contrast to the classification algorithm employed by TeamBeam, none of the Weka algorithms exploit sequence information. In terms of performance, the Bagging meta classifier in combination with a Random Forest base classifier achieves the best overall result, matching the performance of the OpenNLP Maximum Entropy results, but the Bagging/Random Forest combination is associated with a high run-time complexity. The performance of all classification algorithms is relatively close to each other, indicating that the features were well engineered and well suited to the task.
| Algorithm |
Precision |
Recall |
|
| Decision Tree |
0.86 |
0.60 |
|
| Random Forest |
0.94 |
0.87 |
|
| AdaBoost (Random Forest) |
0.94 |
0.91 |
|
| Bagging (Random Forest) |
0.95 |
0.92 |
|
| Naive Bayes |
0.95 |
0.91 |
|
| Bayes Net |
0.93 |
0.93 |
|
| Complementary Naive Bayes |
0.97 |
0.86 |
|
| k-NN (k=1) |
0.90 |
0.93 |
|
| k-NN (k=3) |
0.92 |
0.92 |
|
| SVM (linear) |
0.92 |
0.88 |
|
| Logistic Regression |
0.93 |
0.89 |
|
Table 6: Comparison of various classification algorithms for text blocks, using the PubMed data set, for the Title-Block class. The training set, as well as the test set, contains 1,000 randomly selected articles.
4.4 Token Classification
The evaluation of the token classification is similar to the text block classification. Instead of blocks of text, the individual words within the text are labelled. Again, differences between the three data sets can be observed (Table 7). The training and testing sets are identical to the text block classification evaluation. They each contain 1,000 randomly selected articles. From the data sets, the E-Prints articles appear to be the most diverse set and therefore the performance falls a little bit behind in comparison.
| |
E-Prints |
Mendeley |
PubMed |
| Token Type |
Precision |
Recall |
Precision |
Recall |
Precision |
Recall |
| Given Name |
0.84 |
0.86 |
0.84 |
0.97 |
0.96 |
0.96 |
| Middle Name |
0.84 |
0.90 |
0.90 |
0.90 |
0.94 |
0.91 |
| Surname |
0.86 |
0.87 |
0.84 |
0.93 |
0.94 |
0.95 |
| Index |
0.88 |
0.87 |
0.88 |
0.97 |
0.98 |
0.97 |
| Separator |
0.89 |
0.81 |
0.96 |
0.96 |
0.96 |
0.98 |
| E-Mail |
|
|
|
|
0.96 |
0.98 |
| Affiliation-Start |
|
|
|
|
0.68 |
0.48 |
| Affiliation |
|
|
|
|
0.65 |
0.55 |
Table 7: Evaluation results of the token classification for the three data sets. Precision and recall measure the performance of the classification stage, without any errors which are introduced in previous processing steps.
The PubMed articles provide the best overall performance numbers. Maybe this can be attributed to the additional available classes for e-mail addresses and affiliation information. In many cases a single text block contains the author names, as well as additional information. In such cases the labelling of more classes might improve the individual classification results. From the individual token classes, the initial word of an affiliation appears to be the hardest task, where only less than half of them are successfully identified in the test scenario.
4.5 Meta-Data Extraction
4.5.1 Title Meta-Data
The meta-data Title, Journal and Abstract are taken directly from their respective text blocks. For the Title meta-data the classification phase introduces only a small error. The preceding text extraction and text block identification steps are the main sources for most of the cases where the extraction process failed. To assess the impact of the pre-processing steps, in addition to the overall precision and recall, the theoretical upper limit for recall is also presented (Table 8). The maximum recall value is calculated by computing the fraction of articles where a single text block on the first page contains the correct title.
| Data Set |
Precision |
Recall |
Max. Recall |
|
| E-Prints |
0.87 |
0.70 |
0.75 |
|
| Mendeley |
0.94 |
0.92 |
0.94 |
|
| PubMed |
0.92 |
0.83 |
0.88 |
|
| Mendeley 10k |
0.96 |
0.94 |
0.96 |
|
| PubMed 10k |
0.93 |
0.87 |
0.89 |
|
Table 8: Performance of meta-data extraction with the TeamBeam algorithm for titles over all three data sets. For the last two rows the training set and test set contained 10,000 articles instead of 1,000 as for the other rows.
Similar to the reference algorithms (ParsCit, Layout-CRFs and Mendeley Desktop), the extraction process works best on the Mendeley data set and worst for the E-Prints articles. The performance numbers for the PubMed data set fall in between. Generally the precision and recall values for TeamBeam compare favourably to the reference algorithms and are well above the heuristic baseline of 0.62.
4.5.2 Impact of the Training Set Size
The classification result as well as the language model depend on the number of training articles. The more training examples that are fed into the classification algorithm, the more robust the results are expected to be. At a certain point, the improvements gained by adding more articles should decrease.
To assess the influence of the training set size, an initial split of the PubMed data set was created. This split contains 10,000 training examples and 1,000 test articles. The training set was further sampled to measure the impact of the number of training examples on the title meta-data extraction process (shown in Figure 2).
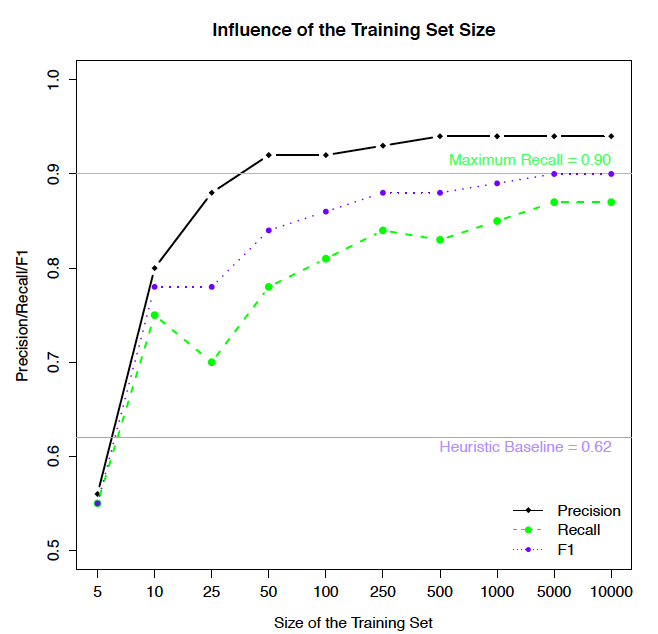 Figure 2: Influence of the number of articles used for training on the title extraction result. The PubMed data set serves as the basis for comparison, where the same 1,000 articles are used for testing all training sets.
Figure 2: Influence of the number of articles used for training on the title extraction result. The PubMed data set serves as the basis for comparison, where the same 1,000 articles are used for testing all training sets.
One can observe that the point of saturation differs between the precision and the recall evaluation metrics. While there are no further gains in the ratio of correctly extracted titles at about 500 articles, the number of retrieved titles reaches its plateau at about 5,000 articles.
4.5.3 Author Meta-Data
Author meta-data extraction is the final stage of the TeamBeam processing pipeline. All errors of the previous stages are accumulated and contribute to the overall results. The numbers reported for the TeamBeam algorithm cannot be directly compared to the results of the reference algorithms, as no distinction is made between the individual parts of an author name.
One can observe a wide variation of performance numbers for the three data sets and the two meta-data types (Table 9). Generally surnames appear to be easier to extract than given names. There are multiple possible reasons for this discrepancy, for example given names are often abbreviated. Furthermore, the distinction between a given name and a middle name is not obvious from the word itself.
| |
Given Name |
Surname |
| Data Set |
Precision |
Recall |
Precision |
Recall |
| E-Prints |
0.53 |
0.36 |
0.86 |
0.68 |
| Mendeley |
0.70 |
0.69 |
0.84 |
0.82 |
| PubMed |
0.90 |
0.84 |
0.92 |
0.87 |
| Mendeley 10k |
0.79 |
0.75 |
0.92 |
0.87 |
| PubMed 10k |
0.90 |
0.88 |
0.92 |
0.90 |
Table 9: Overview of the meta-data extraction for all three data sets for given names and surnames. For the last two rows the training and testing sets contained 10,000 articles.
Increasing the training set does not appear to improve the precision of the extraction, but has a positive effect on the recall. This is expected behaviour, as the classifier is fed with more examples of different author names, which will result in a higher number of recognised names. Given a sufficient number of training articles, the difference between the Mendeley and PubMed data sets reduces, at least for surnames.
5 Discussion
Propagation of Errors: As the evaluation showed, by far the largest contributor to the missing meta-data is not the classification component, but the preceding text extraction and text block identification. For example, there is a single text block that contains the title of the article in only 75% of all articles in the E-Prints test data set. Any gains made in text processing will improve the overall meta-data extraction results.
Availability of Meta-Data: It is not uncommon that the first page is not part of the original article, especially in the E-Prints data set, but some sort of cover page. These pages differ in layout and content from typical article pages. Usually these pages do not contain any meta-data related to the authors, such as their e-mail addresses. For some journals in the medical domain there is only additional author information if the author is the corresponding author. Sometimes this information is presented not on the first page, but on one of the last pages.
Author Meta-Data: Although the classification algorithm takes the sequence into account, the results could be improved by applying some post processing. For instance, one could rule out any cases where a middle name is directly followed by a given name. OpenNLP provides facilities to integrate such heuristics into the computations, but in the current version of TeamBeam these are not used.
Affiliation Meta-Data: The TeamBeam algorithm uses common name lists for given names and surnames, but not for organisations. A list of universities and institutions will certainly help to improve the classification results for the affiliation meta-data as well.
E-Mail Meta-Data: Although e-mail addresses appear to be fairly easy to extract, there are cases which can only be resolved via additional logic. For example, in the area of computer science multiple e-mail addresses with the same domain name will often be abbreviated to: {name1, name2}@domain.tld.
Selection of Training Set: The selection of the training data set has a big impact on the final performance of the meta-data extraction process. If certain layouts or formatting appear infrequently in the training set, it will result in poor extraction quality. For example, the ACM layout appears to be sufficiently different from the majority of articles present in the Mendeley and PubMed data sets. Creating dedicated training sets for specific layout types might help to improve the extraction performance. Furthermore, it has been observed that for small training sets the performance varies considerably, which indicates that even with a small but carefully selected training data set, good results can be achieved.
6 Conclusion
The TeamBeam meta-data extraction algorithm builds upon a text extraction component that parses scientific articles stored as PDF files and identifies blocks of text based on layout and formatting information. For the meta-data extraction, these text blocks are first classified into a set of categories. In some cases, these categories directly correspond to a meta-data field such as a title or the name of a journal.
For meta-data which relates to the author, for example the name, e-mail address and affiliation, the output of the first stage is fed into a second classification phase. The individual words within the text blocks are classified. The output is a rich set of meta-data, which describes the scientific articles and their authors.
An extensive evaluation was conducted, based on three different data sets, where two of them are publicly available. First, a set of baselines were defined using simple heuristics, which turned out to perform reasonably well. Next, three established meta-data extraction approaches were presented with results on two of the data sets used. During the evaluation, the individual components of the TeamBeam algorithm were analysed. It was found that the approach introduced does deliver a satisfactory level of performance for all data sets.
The algorithm can be further improved, especially in terms of applied heuristics which are in part already used by alternative approaches. The text processing components, which are applied prior to the meta-data extraction task, also limit the algorithm's performance. Both areas will be addressed in future work, as well as extending the extraction to an even richer set of meta-data, to include, for example, the references.
7 Acknowledgements
This work was funded by the European Commission as part of the TEAM IAPP project (grant no. 251514) within the FP7 People Programme (Marie Curie).
8 References
[1] A. L. Berger. A Maximum Entropy Approach to Natural Language Processing. Computational Linguistics, pages 1—36, 1996.
[2] I. G. Councill, C. L. Giles, and M.-y. Kan. ParsCit: An open-source CRF Reference String Parsing Package. In N. Calzolari, K. Choukri, B. Maegaard, J. Mariani, J. Odjik, S. Piperidis, and D. Tapias, editors, Proceedings of LREC, volume 2008, pages 661—667. Citeseer, European Language Resources Association (ELRA), 2008.
[3] M. Granitzer, M. Hristakeva, R. Knight, K. Jack, and R. Kern. A Comparison of Layout based Bibliographic Metadata Extraction Techniques. In WIMS'12 — International Conference on Web Intelligence, Mining and Semantics, page to appear. ACM New York, NY, USA, 2012.
[4] H. Han, C. L. Giles, E. Manavoglu, H. Zha, Z. Zhang, and E. A. Fox. Automatic document metadata
extraction using support vector machines. 2003 Joint Conference on Digital Libraries 2003 Proceedings, pages 37—48, 2003.
[5] H. Han, E. Manavoglu, H. Zha, K. Tsioutsiouliklis, C. L. Giles, and X. Zhang. Rule-based word clustering for document metadata extraction. Proceedings of the 2005 ACM symposium on Applied computing SAC 05,
page 1049, 2005.
[6] M.-t. Luong, T. D. Nguyen, and M.-y. Kan. Logical Structure Recovery in Scholarly Articles with Rich
Document Features. International Journal of Digital Library Systems, 1(4):1—23, Jan. 2010.
[7] C. D. Manning, P. Raghavan, and H. Schütze. Introduction to Information Retrieval, volume 61. Cambridge University Press, 2008.
[8] A. Ratnaparkhi. Maximum Entropy Models for Natural Langual Ambiguity Resolution. PhD thesis, 1998.
[9] K. Seymore, A. McCallum, and R. Rosenfeld. Learning Hidden Markov Model Structure for Information Extraction. In Science, pages 37—42. In AAAI 99 Workshop on Machine Learning for Information Extraction, 1999.
About the Authors
 |
Roman Kern is senior researcher at the Graz University of Technology, where he works on information retrieval, social web and natural language processing. In addition he has a strong background in machine learning, which he acquired during his work at the Know-Center. Previous to working in research he gained experience in industry projects at Hyperwave and DaimlerChrysler. There he worked as project manager, software architect and software engineer for a couple of years. He obtained his Master's degree (DI) in Telematics at the Technical University of Graz. He published over 20 peer-reviewed publications and achieved top rank results in international scientific challenges like CLEF, ACL SemEval. Most of his publication work can be accessed online: http://www.mendeley.com/profiles/roman-kern/.
|
 |
Kris Jack is the Chief Data Scientist at Mendeley and is responsible for the development of their data mining technologies. He has over ten years of experience in both academia (PhD in AI, University of Dundee; Research Associate, NaCTeM, UK) and industry (Expert R&D Engineer in Orange Labs and the CEA) of solving complex large scale data problems.
|
 |
Maya Hristakeva is a Research Scientist with Cognitive Match Ltd. Prior to this, she was a Data Mining Engineer at Mendeley Ltd, where she worked on information extraction, text classification, recommender systems, as well as crowdsourcing Mendeley's extensive catalogue of bibliographic data. Maya has also worked at C8 Medisensors as a Researcher focused on developing algorithms for continuous non-invasive
measurement of blood sugar in humans. Maya Hristakeva holds a Master of Science degree in Machine Learning from University of California — Santa Cruz. Her current research interests are focused on applying machine learning techniques for behavioral targeting.
|
 |
Michael Granitzer is Professor for Media Informatics at the University of Passau, Germany. Before, he was Scientific Director of the Know-Center Graz and Assistant Professor at the Knowledge Management Institute of Graz University of Technology, where he worked on topics like automatic text classification, large scale text retrieval and information visualisation. As a Marie Curie Research Fellow at Mendeley Ltd., Prof. Dr. Granitzer investigated machine learning and information retrieval techniques for managing large academic knowledge bases. Dr. Granitzer published around 110 mostly peer-reviewed publications including journal publications, book chapters and books in the fields of information retrieval, machine learning, knowledge discovery, text mining and visual analytics. He has been a member of several program committees and a reviewer for well known journals such as Elsevier's Pattern Recognition, Springer's Journal on Multimedia Tools and Applications and the IEEE Journal on Transactions of Knowledge and Data Engineering. As principal investigator, he managed several nationally and internationally funded research projects including the 4 times awarded FIT-IT project DYONIPOS (DYnamic ONtology based Integrated Process OptimiSation), the Marie Curie FP 7 IAPP Project TEAM (Transferring knowledgE in Academic knowl- edge), the Marie Curie FP 7 IRSES project WIQ-EI (Web Information Quality Evaluation Initiative) and the FP 7 STREP CODE Commercially empowered Linked Open Data Ecosystems in Research.
|
|