D-Lib Magazine
January/February 2017
Volume 23, Number 1/2
Table of Contents
Opening the Publication Process with Executable Research Compendia
Daniel Nüst*
Institute for Geoinformatics, Münster
daniel.nuest [at] uni-muenster.de
Markus Konkol*
Institute for Geoinformatics, Münster
m.konkol [at] uni-muenster.de
Marc Schutzeichel
University and State Library, Münster
m.schutzeichel [at] uni-muenster.de
Edzer Pebesma
Institute for Geoinformatics, Münster
edzer.pebesma [at] uni-muenster.de
Christian Kray
Institute for Geoinformatics, Münster
c.kray [at] uni-muenster.de
Holger Przibytzin
University and State Library, Münster
holger.przibytzin [at] uni-muenster.de
Jörg Lorenz
University and State Library, Münster
joerg.lorenz [at] uni-muenster.de
*Shared co-first authorship
https://doi.org/10.1045/january2017-nuest
Abstract
A strong movement towards openness has seized science. Open data and methods, open source software, Open Access, open reviews, and open research platforms provide the legal and technical solutions to new forms of research and publishing. However, publishing reproducible research is still not common practice. Reasons include a lack of incentives and a missing standardized infrastructure for providing research material such as data sets and source code together with a scientific paper. Therefore we first study fundamentals and existing approaches. On that basis, our key contributions are the identification of core requirements of authors, readers, publishers, curators, as well as preservationists and the subsequent description of an executable research compendium (ERC). It is the main component of a publication process providing a new way to publish and access computational research. ERCs provide a new standardisable packaging mechanism which combines data, software, text, and a user interface description. We discuss the potential of ERCs and their challenges in the context of user requirements and the established publication processes. We conclude that ERCs provide a novel potential to find, explore, reuse, and archive computer-based research.
Keywords: Executable Research Compendium, ERC, Open Access, Containerization, Research Data, Computational Research
1 Introduction
Open Access is not only a form of publishing that ensures research papers become available to the large public free of charge, it is also connected to a trend towards Open Science, which makes research more transparent (Nosek et al., 2015, and see also Charles W. Bailey, What is Open Access? and Open Access to
Scientific Information, History of the Open Access Movement). To fully realise Open [computational] Science we expect everyone can reproduce findings because access is granted to papers, research data, methodology, and the computational environment. In parallel, the scientific paper "is evolving into a multi-part distributed object that can include an article, data, software, and more" (Hanson et al., 2015). Access to these components is rarely given, making it challenging to archive and reproduce methods and results. Reasons include privacy concerns and copyright issues. Another key obstacle is the lack of standardized means for sharing all the parts of an 'evolved' scientific paper in an easy way (Hanson et al., 2015).
The main contribution of this work is the definition of a compendium-based publication process. It facilitates Open Science and enables new ways to conduct research. Its core component is the executable research compendium (ERC), which opens and integrates the scientific process across all activities and stakeholders. The compendium includes, besides the actual paper, source code, the computational environment, the data set, and a definition of a user interface. It has internal connections which facilitate new ways of interacting and reuse. Such a compendium is a self-contained entity and can be executed almost entirely on its own, requiring only a generic virtualization environment. ERCs are intended for the huge number of research projects which live on a researcher's desktop computer. Larger scale undertakings with high computational or storage requirements, e.g. distributed infrastructures, and using external third-party services ("black boxes") are out of scope of this work although being subject to vivid research (cf. Chen et al., 2016, Goecks et al., 2010).
The paper is structured as follows. The next section provides a review of related work and basic concepts. In the main part, we explain how the ERC builds upon stakeholder requirements and current technologies. We describe the process of publishing ERCs and conclude by highlighting the main contributions and future work.
2 Related Work
In this chapter we introduce the term "reproducibility" and Open Access as motivators of an open publication process. Our work builds on the concept of research compendia/objects, data platforms, and methods to capture software environments. We also summarize key challenges in open reproducible research.
2.1 Definition of Reproducibility
In current research, talking about computational reproducibility can lead to confusion as a universally accepted definition of the term "reproducibility" is not yet established. While some researchers treat "reproducibility" and "replicability" as interchangeable terms (Bollen et al., 2015), others distinguish them carefully (Leek and Peng, 2015; Goodman et al., 2016). "Replicability" is given if researchers receive the same results by using the same methodology but a different data set (Bollen et al., 2015). Research findings are "reproducible" if researchers are able to compute the same results by using the same procedure and the same data set (Bollen et al., 2015). This means a researcher requires the entire set of information which constitute the basis of the results reported in the paper including data sets, source code, and configuration details (Vandewalle et al., 2009). A key difference between the two terms is the data set which needs to be different from the originally used data set in the case of "replicability", and to be the same in the case of "reproducibility". Consequently, it does not mean reproducible research findings are true. They can still be subject to flaws in the study design (Leek and Peng, 2015). However, reproducible outcomes are more reliable as they allow other researchers to understand the (computational) steps described in the paper. In open reproducible research, the components required to reproduce the results are publicly available. Other scientists are thus able to reuse parts for their own research.
2.2 Open Data & Open Access
Today, most researchers find it difficult to reproduce the analysis reported in papers published 5-10 years ago, or even recover the data. For instance, Vines et al. (2014) reported a half-life of four years for data recovery from the original authors. Nowadays there is a strong trend towards publishing scientific papers as Open Access, meaning anyone has free access to the published material, including free access to data and software (cf. The Open Definition: "Open data and content can be freely used, modified, and shared by anyone for any purpose."). Different models exist (cf. Harnad et al., 2004) and publishers increasingly offer Open Access routes for research papers. For instance, PLoS-One's policy requires authors to publish data by default (Bloom et al., 2014). The Directory of Open Access Journals (DOAJ) lists over 9000 journals. Open Access is a key topic in the European Union's research programme Horizon 2020. Dissemination and reuse of scientific findings shall be improved by making them accessible at no charge through suitable e-infrastructures (European Commission, 2015) such as the European Open Science Cloud.
2.3 Research Compendia & Research Objects
Gentleman and Temple Lang (2007) use the term Research Compendium (RC) to refer to the unit of scholarly communication which includes the research paper, the code, and the data with a "dynamic document" (cf. Knuth, 1984) at the core. They present the advantages and potential uses of (executable) research compendia, which allow to completely reproduce the computational aspects of a scientific paper. (Randomness of specific simulations or machine learning methods can be handled by setting a seed. Numerical differences between runs can be mitigated by appropriate rounding. Both these aspects require careful design of the actual analysis and are beyond the scope of ERC.) The practical implementation focuses on "single language compendia", which are essentially the respective languages' standard packaging mechanism (e.g. R packages). Stodden et al. (2015) developed a platform for sharing, executing and validating Research Compendia.
Research Objects (RO) are "semantically rich aggregations of resources that bring together the data, methods and people involved in (scientific) investigations" (Bechhofer et al., 2013). They comprise metadata standards and bundling of any kind of resource across a range of scopes, such as scientific workflows (Belhajjame et al., 2015), preservation (Research Object BagIt archive), or computational jobs. These can be included or remote (i.e. linked) resources. ROs are also Linked Data resources themselves and can be managed and preserved in a tailored platform (Palma et al., 2013).
Both Nature and Nature Geosciences published about the need to publish reproducible code (Nature Editorial, 2014; Easterbrook, 2014). The majority of scientific journals allow adding supplementary material to publications, and hence creating and publishing Research Compendia or Research Objects has been possible for a long time. Yet current supplemental material frequently looks very much like a directory on the researcher's personal computer.
2.4 Research Data Platforms
We see a large number of technical solutions for research repositories, including RunMyCode, the Open Science Framework, Zenodo, and figshare (see Dave Wilkinson's "rubric" for platforms.) All these platforms provide identification (some only by email; others support ORCID or GitHub identities), uploading, and persistent identifiers for citation of deposited items. All platforms have a data size limitation, some have paid upgrade plans. Some sites argue users should trust their long-term availability, others just offer the service as-is, for example, RunMyCode is provided on an "as is" and "as available" basis. All these platforms are fine for viewing and downloading, but none of them actually performs the computations needed to carry out reproducible research. This is most likely due to recomputation requiring considerably more resources than storage, and thus causing substantial costs, and security concerns. Also, the platform would need to understand a plethora of different computational setups and would have to cater for execution of compendia within these.
2.5 Capture Workflows and Runtime Environments
Researchers enjoy to conduct their research in an environment composed of software of their own choice. Consequently, the number of unique research environments approximates the number of researchers, and hence expecting one standardised computational workflow would not be feasible. But the complete computational environment of the original researcher, the runtime environment, is an important aspect of reproducibility. Different approaches to capture the workflows and runtimes can be chosen.
Regarding workflows, a commonality of all computational sciences is carrying out a number of steps (process input file(s), process data into result, e.g. number or graph). These steps are ordered and must be documented to allow understanding, execution, and reproduction. The canonical example is the classic unix utility "make" (Mecklenburg, 2004), which executes commands according to the instructions in a Makefile. The concept has been adapted for specific scenarios, e.g. remake for data analysis in R, drake for data workflow management, or Taverna (Wolstencroft et al., 2013) for web-based workflows. Santana-Perez et al. (2016) demonstrate a semantic modelling approach to conserve scientific workflow executions based on semantic vocabularies. Thain et al. (2015) discuss two broad approaches to capture scientific software executions: "preserving the mess, and encouraging cleanliness". They provide an extensive picture on technical and organizational challenges in the context of in-silico experiments and present prototypical solutions (e.g. Umbrella).
Alternatively, the environment can be confined to an interpreter of a particular language, such as R (R Core Team, 2014). The environment is reproduced if the correct versions of the interpreter and all extension packages/libraries/modules are used. These dependencies quickly become complex so a manual documentation is not feasible. The R extension packages checkpoint or packrat ease the process of reproducing project dependencies, except the interpreter itself. ReproZip (Chirigati et al., 2013) or Parrot (Thain et al., 2015) apply tracing techniques to capture the minimal set of objects and commands needed for an analysis and use it to build a virtual machine (VM) or container image for execution.
Containerization originates from packaging applications and their dependencies for deployment in cloud infrastructures (cf. Dua et al., 2014). Containers have proven to be a suitable technology in reproducible research: Howe (2012) lists improvements virtualization and cloud computing provide for reproducibility, all of which apply directly to containerization. Boettiger (2015) demonstrates its usage for computational analysis in R and derives best practices. Marwick (2015) accompanies a research article by Clarkson et al. (2015) with a complete research compendium containing the analysis (also available on GitHub). Hung et al. (2016) use containers to package graphical user interface-based research environments with multiple tools from multiple languages across operating systems in the context of bioinformatics.
The dominant platform for container is Docker (Merkel, 2014). Its images are created using a recipe called Dockerfile — a Makefile with inheritance support for creating containers. Images can be build and executed on any machine running a Docker host, and are distributed in ready to run form via online hubs, for example Docker Hub or Quay.
2.6 Challenges in Open Reproducible Research
Solutions to technical and legal challenges with regard to implementing reproducible research exist (cf. Stodden et al., 2014). Other challenges, such as the question of incentives and developing reward mechanisms, are closely connected to the intricacies of academic authorship and credit. The challenges and some approaches are summarized in the Wikipedia article on academic authorship, for example the individual h-index (George A. Lozano) or Nature's author contributions statements.
Among the reasons why researchers do not publish reproducibly, Borgman (2007) mentions (1) a lack of incentives in terms of citations or promotion, (2) the effort required to clean data and codes, (3) the creation of a competitive advantage over other fellows, and (4) intellectual property issues. Other reasons include privacy or confidentiality issues (Glandon, 2012). None of these issues can be solved by technical solutions alone. Instead they require a discourse within the scientific community, or even a mindshift to "build a 'culture of reproducibility'" based on selfish reasons to publish reproducibly (Markowetz, 2015).
Although moving the relevant parts of a researcher's hard drive to public and citable archives is a step forward in the direction of Open Access to research findings, we believe this alone will not convince researchers to adopt it as standard practice. Such archived workspaces could be incomprehensible (Easterbrook, 2014). The potential reward is considered smaller than the possible reputation damage or expected follow-up work, e.g. support questions.
Each scientific community must address such worries to make publication of workspaces an effective incentive for reproduction. Encouraging statements such as Nick Barnes' column "Publish your computer code: it is good enough" (Barnes, 2010) pave the way towards venturing reproducibility of software. However, they also highlight the necessity for communication of researchers' uncertainties regarding reproducibility as a deliberate act of exposure to critics and competitors.
The research data repositories mentioned before support reproducibility, but they cannot carry out the actual reproduction, nor do they suggest conventions for doing so in an automated fashion. Although they are designed to deposit code and data supplementing a scientific paper, none of them require documenting the runtime environment, under which the reproduction material is expected to reproduce the paper, in a systematic way. This puts more burden on the original author, as each is supposed to know how to describe this in a generic way. The same burden is placed on a reader as the reproducer.
3 The Executable Research Compendium
3.1 Stakeholders and Roles in the Publication Process
The ERC brings together perspectives of different user groups involved in scientific publications. To develop a common ground for them, we first consider some key requirements of users separately. We identified these requirements in discussions including experts from university, library, and publishing (members of the project team and external partners are listed here).
Authors must be given support to create an ERC without too much additional work, ideally starting from their digital workspace "as is". Since filling out forms for metadata is often perceived as a daunting and time-consuming task, an ERC should be restricted to a minimal set of required metadata. Ideally, the needed information can be (semi-)automatically deduced via existing identifiers and catalogues, such as DataCite (Brase, 2009) for data sets and ORCID (Haak et al., 2012) for researchers, thus avoiding the need for entering them multiple times. If common practices for reproducible research are followed then creating an ERC should be as simple as manually editing a configuration file and running a single command.
Readers want to be able to rerun the analysis in an ERC. On a basic level, a compendium must provide a simple "start" button to run an analysis and a traffic-light like signal to learn about the result, cf. Elsevier's Earth and Planetary Innovation Challenge winning submission "One-Click-Reproduce" by Edzer Pebesma. A green light shows a successful, a red one an unsuccessful reproduction of the results. Here a green light means the computational steps are executable and the results are equal to those submitted by the original author. A red light means this is not the case. Either way the status light is no indication on the validity of the findings. At an advanced level, readers can examine detailed information on, for example, data and code underlying figures, or errors preventing successful execution. A typical reader's question might be about assumptions and concrete tools the authors did not mention. Moreover, it would be interesting for readers to see how the results change and if the conclusions still hold true after manipulating parameters.
Reviewers must be put in the position to scrutinize a piece of research. While potential malicious intent of the submitting authors is not considered here, review processes still involve a trust component as far as scientific and scholarly best practices are concerned. Reviewers often have to rely on the analysis in a research paper to be complete, correct, and consistent with the included visualizations. In order to verify the analysis, they need tools to recreate computation-based analyses with minimal effort. As a review and publication process is well established, the ERC shall align itself with it. It becomes the item under review and should help to increase the quality of the review. Since this also increases the quality of journals, ERCs ultimately serve the needs of editors, authors, readers, and publishers.
Libraries and publishers make research articles accessible for the scientific community and the public. However, access to the source material is not always granted. One of the reasons for this is neither libraries nor researchers are currently familiar with a publication process that allows for including procedures used to produce research outcomes (tables, figures). Any new tool for this purpose must leverage their expertise and pay attention to their existing workflows.
Curators and preservationists require the reader's basic level to assess a compendium's status. A compendium run must entail an execution of the contained computation and an automatic validation of the result to ensure the integrity of the digital asset. Since they need to integrate an ERC into a curation workflow, the required metadata must follow standards for digital preservation. As they cannot rely on the execution platform to be persistent, they need access to relevant metadata in "their" formats independently from the data and software within the compendium. Subsequently, the top-level package of the compendium should follow a preservation standard.
To fulfil the needs of these different user groups, we define conventions in the remainder of this chapter. They allow to create research compendia supporting automatic execution, and to create services for executing such research compendia.
3.2 Core Parts of ERC
The core parts of an ERC are data, software, documentation, and user interface (UI) bindings (see Figure 1).
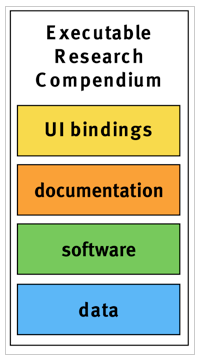
Figure 1: The core parts of an ERC are data, software, documentation, and UI bindings.
Data comprises all inputs for an analysis, ideally starting with raw measurements, for example, in form of text files, or databases.
Software comprises code created by a researcher and all underlying libraries or tools to reproduce the analysis in form of scripts/source code, a Dockerfile, and a Docker container.
Documentation comprises both instructions, such as a README file, and the actual scientific publication, e.g. in PDF format, any supplemental records, and metadata in standardized formats. The actual publication is the main output of the compendium and the core element for validation. An important metadata element are licenses for the different parts of a compendium.
UI bindings provide linkage between research components and user interface widgets. They can be used to attach UI widgets to static diagrams in order to make them interactive. Their representation can be stored as metadata within an ERC as part of the documentation. The resulting UI widgets open up the container and allow readers to drill deeper into results. UI bindings can unveil parameters which are required for a comprehensive understanding but are often buried in the code.
3.3 Creation and Reproducibility
Two approaches for creating an ERC are possible: post-hoc or on-the-fly.
Post-hoc creation is based on the regular workspace, which is a collection of files (data, code, documentation). Authors of a publication can submit a workspace to an ERC building service to generate a compendium. Such a service requires complex logic and user intervention, either by an expert on the service provider side or by the author, to detect how to start the analysis and how to validate its result. Following best practices from reproducible research, such as literate programming (Knuth, 1984) or having a default main file and execution command (comparable to a Makefile), can considerably reduce the required intervention. An ERC specification can support this with a "convention over configuration" approach (see Wikipedia, Convention over configuration, Nicholas Chen, "Convention over Configuration" and also Maven standard directory layout). Information that cannot be derived automatically must be elicited during the submission process, ideally with pre-filled forms.
Alternatively the creation and maintenance of a compendium happens on-the-fly while carrying out research. A user starts with a template compendium. Since the compendium evolves with the work in progress, it can be executed and checked regularly.
In both cases, the ERC is bound to a specific publication document. This "exit point" is also well-defined in the ERC and is exploited for result validation and to highlight changes in the results after parameter manipulation.
In order to reproduce an analysis, we make use of the containerization technology Docker. Storing both a docker image and the corresponding Dockerfile creates two levels of reproduction: (i) the ability to provide the original runtime environment, and (ii) recreating it from scratch, potentially in a more recent version. Docker mitigates some issues for the reproduction service to run the software of the original researcher, for example with a varying environment of the workspace (Linux, Windows, OS-X), but cannot solve others, for example requiring licensed software (MATLAB™, ArcGIS™) which must not be redistributed.
Existing approaches using Docker to replicate environments (see previous chapter) require readers to follow individual written instructions, and to have expertise in the used software setup. This is not apt for a one-click execution (for readers) nor for automatic content validation (for reviewers and curators). The ERC defines machine-readable conventions for computer systems to control and evaluate the embedded container, namely command-line interface instructions to run it (e.g. "docker run" as part of ERC metadata), and rules to check a successful execution based on the created workspace. The Dockerfile used to build this container defines the environment and command for analysis execution.
After an ERC's execution, the result is evaluated. A minimal evaluation relies on the exit code of the main process in the container. At an advanced level, checksums or contents of files and execution logs can be evaluated.
Once the analysis can be executed by the creation service, the information accessible at runtime, e.g. actually loaded modules and libraries as well as dataset objects, is a fast and reliable way to derive both software metadata and dataset metadata. The potential information stretches from detailed names and versions of attached libraries to the spatial and temporal extent of the data subsets. This is prone to be more correct than analysing source code, using system-wide installed software, trying to read all imaginable data formats while assuming all data files in a workspace are actually used, or manual documentation.
Docker is well suited for storing and transporting software and its dependencies, but it does not serve the needs of data repositories and archives well, that are concerned with bitstream preservation and integrity of files.
3.4 Publication Process
An ERC facilitates a scientific publication that is not only composed of a paper but also the data, the source code, and the software environment, packaged in an executable manner. The executable research compendium described in this work is a fundamental component of the proposed publication process consisting of four consecutive steps (see Figure 2). The process is aligned with peer-review based journals. Their existing practices and protocols can be applied for transfer of compendia between stages and handling the required data, such as related communications or the state within the process.
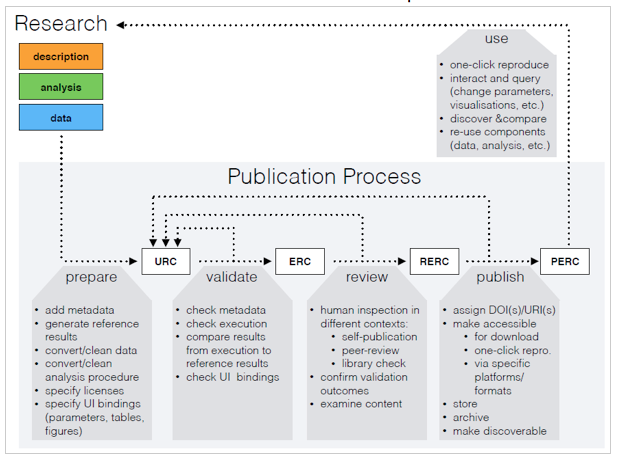
Figure 2: The ERC-based publication process: a research workspace is prepared for a URC, which is validated to become an ERC, which turns into an RERC after peer review, and eventually published as a PERC to be used, e.g. for subsequent cycles.
In the first subprocess, the author prepares the research material as described in the previous section resulting in an unvalidated research compendium (URC). A URC is neither executable, nor necessarily visible to the public, but it ideally contains already the entire research material including data set, source code, and the software environment. A URC forms the input to the validation subprocess which involves (i) metadata verification, (ii) the error-free execution of the reproducible parts of the paper, and (iii) a confirmation from the primary author that the automatically reproduced results are identical to the actual publication. The first two aspects should be automated as part of a submission. If the validation succeeds, it produces an executable research compendium (ERC), which is passed on to the next subprocess.
The review process focuses on human inspection. It is necessary to avoid publishing meaningless or questionable compendia. Human inspection can be done by library staff (e.g. to ensure adherence to non-formal criteria), maintainers of a data repository (e.g. to filter offending or illegal content), journal editors and peer reviewers (e.g. to warrant scientific quality in the context of a journal submission). If the review process has a positive outcome, it produces a reviewed executable research compendium (RERC), which constitutes the input to the final subprocess: publish. During this subprocess, a RERC is enriched (e.g. by assigning DOIs/URIs) and then made accessible as a published executable research compendium (PERC). Researchers can reproduce the results reported in a PERC via a single click. They can also reuse components of a PERC (e.g. its data) for their own research. In order to make PERCs discoverable and comparable, there has to be a minimum of meta information attached to them.
Finally, if the authors provide UI bindings during the publication process, readers can interact with an ERC in a much deeper way. For example, if the authors specify which variables in the code contain specific threshold values, then readers of the PERC can interactively change them — i.e. via an automatically generated UI — to explore whether the reported outcomes still hold true for different threshold values. Another benefit relates to diagrams and visualizations: readers could select from different visualizations to more easily compare it to a diagram in a second paper.
3.5 Findability and Preservation
ERCs can have layers of meta information which are divided into reproducibility metadata and discovery metadata.
Packages, libraries, and specific versions of software is crucial information for reproducibility. Therefore these must be made available for the URC's validation.
The ERC itself is conceptualized as a ready-for-ingest digital asset as defined in OAIS reference model (CCSDS, 2012). Its outer container uses the BagIt file packaging format by Kunze et al. (2011). It provides minimal metadata and checksums for a file based payload. The payload comprises at least (i) the aforementioned Docker image (an inner container) and Dockerfile, (ii) metadata files in different formats such as Codemeta, DataCite (Brase, 2009), (iii) the workspace of the conducted research including code and data files, and (iv) created output documents, e.g. a PDF of the original paper. Research data and code can be stored in the image to make distributed reproduction easier but making them accessible as part of the outer container is advantageous for long-term preservation. This is also the case for the metadata.
By design ERCs encapsulate any information relevant to the reproduction of its contents (cf. "Core parts of ERC"). External contextualization of an ERC can be achieved by generating additional discovery metadata and ultimately connecting the ERC as a whole to the Linked Open Data cloud (Bizer et al., 2009) by using discipline-specific vocabularies such as GeoSPARQL. This is done to support findability of the ERC. Semantic references make ERC metadata interoperable and ready for discovery.
For the retrieval of software metadata, it is necessary to identify software dependencies. The metadata included in an ERC also provides structural information needed to connect the entry point for an execution with the internal structure of the underlying parts (code, data, text, UI bindings). It is therefore reasonable to start connecting the available information about the underlying parts as early in the workflow as possible and thus contextualize the files within an ERC.
Our specification also considers preservation requirements by extending bitstream preservation. The recreation of the original computational environment is vital when rerunning the code of a paper. Jon Claerbout's idea of an article being a mere advertising of the underlying scholarship has been concisely paraphrased in Buckheit et al. (1995, p. 5): "The actual scholarship is the complete software development environment and the complete set of instructions which generated the figures". Consequently, when it comes to preservation, this complete environment must be included and the instructions must be executable. This refers to the internal structure and the dependencies.
In addition, ERCs can be integrated into the existing ecosystem of persistent identifiers using mechanisms to uniquely and permanently identify the involved resources and agents. Furthermore, metadata comprises explicit license information, ideally using well-known abbreviations or vocabularies, for each component of a compendium. Authors must be assisted in defining data, software, and text licenses, as this is a tedious but crucial aspect of openness — only a suitable license clearly allows and defines possible reuse.
The user interface definitions (UI bindings) are an optional element of an ERC and allow for interaction with the included analysis. On top of that, they document the analysis code by providing a guide to the main functions and their manipulable parameters. We see different approaches to generate them ranging from fully automatic to manual definition. Fully automatic UI bindings analyze the code and generate UI widgets based on the input type (text, number, etc.). UI bindings are created by the author, for example by means of adding statements in the source code or by using a software designed for this purpose.
3.6 Interaction, Exploration, and Reuse
An ERC not only enables third parties to reproduce the original research results (figures, tables), but also facilitates interaction with them. Because an ERC transparently abstracts diverse analyses, i.e. it is a "white box", it seems trivial to build a web service which downloads a research compendium from a repository, unpacks it, executes it, and returns the results created. This extends the minimalistic control functionalities described before with relevant features. Once reproducibility becomes easy to use via ERCs, scientists can benefit from the ability to drill deeper into the computational instructions. For example, they can interactively investigate whether the originally reported results change if they manipulate an underlying parameter, which are fixed in a 'classical' article. In an ERC they can be exposed as changeable variables, e.g. via dedicated components in the user interfaces such as a slider. The original author could provide information about what constitutes a reasonable range of values for this variable. In addition, being able to inspect analysis procedures more closely can help in detecting errors and in safeguarding the integrity of the publication process. For example, a reader might update an underlying library with known bugs but leave the ERC otherwise unchanged.
The recent Reinhart and Rogoff case (Reinhart and Rogoff, 2010; Herndon et al., 2014) showed how damaging it can be when a paper insufficiently describes the analysis on which its conclusions are based.
4 Discussion
4.1 Approach
In this paper we propose the executable research compendium as a new form for supporting the creation and provision of research results. The four parts of an ERC open up archival of and interaction with computational research with the following improvements: (i) reviewers obtain tools for easily validating results in scientific publications submitted by researchers, (ii) results are well-grounded since the research steps and data described in an article are shared, and (iii) other researchers benefit because they obtain tools for accessing, reusing, and extending research components.
The problem of scientists each having an individual workflow is tackled by packaging not just the workspace with data and code, but also the actual publication, the runtime environment, and UI bindings. ERCs not only enable third parties to reproduce the original research and hence recreate the original research results (figures, tables), but also facilitate interaction and recombination, e.g. with other data or new methods. This recombination is complex and probably feasible only for compatible data respectively computational methods and thus restricted to particular domains.
4.2 Distinction of ERC
ERCs rely on the concept introduced as research compendium (RC) by Gentleman and Temple Lang (2007). The core difference to an RC is that an ERC is aware of its complete software environment and contains the so called "transformation software". The transformation software generates different views (e.g. PDF documents or graphics) from the RC by processing code chunks within dynamic documents, e.g. by passing them to an interpreter of the so called "definition language" (Gentleman and Temple Lang, 2007). The authors mention the possibility to include "general purpose software" as part of the "auxiliary software" into an RC in case a version must be specific or might disappear. ERCs take this one step further and embed all software required to run the compendium's code, including the interpreter of the "definition language". It effectively removes all requirements towards the host machine except a Docker runtime.
Research Objects focus in the linked data technology. As such, they are characterized by aggregation and referencing of distributed resources, including workflows and their execution. In contrast, ERCs seek to provide consistent packaging for simple reproduction ("one-click") and interlinking within the contained parts. Both ROs and ERCs are containers for research data and code needed to preserve and reproduce an analysis but they approach this goal from different directions (cf. RO bundle). ROs focus on the outer perspective: provenance, dependency, interconnections. ERCs target the inner scope: consistency, completeness, independence.
4.3 Creation and Usage
The two creation patterns support the majority of user workflows. Post-hoc is less intrusive during research, while a template can put good reproducible research practices (e.g. literate programming) into effect. It remains to be examined which approach requires less effort or finds higher uptake by researchers, and if the expected benefits of publishing an ERC instead of a classic paper outweigh the additional efforts for authors. In any case researchers have to adjust their workflows to remove all "manual" tasks in favour of replicable scripts.
The packaging format specified by the ERC assists different applications (e.g. one-click reproduce, long-term archival, research information systems) and thus allows future usage independent from the described purposes. The runtime packaging feature is based on open source software with high uptake in industry, but although an open specification process is underway (cf. the Open Container Initiative), longevity issues cannot be put aside at this point.
Storing the image and its recipe, i.e. the Dockerfile, increases the chances of reproducing work long after the original publication. With this burden on the Dockerfile, it must be evaluated what criteria it shall comply with for long-term archival, because clarity and extensiveness (e.g. explicit versions) outweigh typical concerns (e.g. image size, up-to-dateness). Nevertheless, ERCs mitigate some of Howe's (2012) challenges for virtualization, e.g. reuse and limitations to interactivity.
ERCs do not cover privacy issues, requiring to anonymize data prior to publication. This restriction holds for other publication forms as well. ERCs cover security concerns, because the Docker container provides an effective sandboxing mechanism.
Interactivity is confined by computation time. Because manipulation of one parameter requires a rerun of the whole container, an execution platform needs to be transparent and reliable in communicating this issue to the user, for example, by indicating the expected run duration.
The efforts needed to create an ERC can be minimized by using automated metadata derivation. This aspect also applies to the generation of UI bindings, which must be possible without too much effort by the author. We still have to evaluate to which extent UI bindings can be generated automatically.
4.4 Challenges
The key challenges for a publishing process based on ERC are (i) the creation of ERCs must be easy for authors, (ii) ERC-based interaction, discovery, exploration, and reuse must provide sufficient benefits for scientists to result in a broad uptake of the concept, and (iii) ERCs must handle diverse workspaces and integrate requirements from all stakeholders. The adoption of the ERC will be limited if tackling these challenges leads to a system that is too complex or not understood by users.
Some core aspects of the publication process cannot be defined to the required level of detail at this point, namely management of review state (Put review metadata and state into the container or keep it outside?) or transfer and storage (Can journal platforms handle ERC file sizes and execution?). It is inevitable to accompany the concepts in this work by a practical implementation to settle these questions.
Research compendia are designed to support science during preparation, implementation, and publication. However, the definition of ERCs alone cannot enforce correct methodology or proper reviews. Communities of practice have to develop conventions and to expand education to put compendia into effect. Currently, researchers use libraries rarely to curate their work. ERCs can connect the research and library communities as a step towards better digital curation, one of the major challenges for memory institutions of the future.
5 Conclusion
Reproducible research is a goal with extraordinary meaning for scientific publications. ERCs provide an innovation for the publication process by opening its result for reuse. Subsequently they help to implement the goals of Open Science. In this paper we provide the following key contributions:
- a compendium-based publication process
- reproduction of the computational steps, the results, and visualizations in ERCs
- packaging computational research for long-term reproducibility
- new ways of interaction with research
Reproducible research can only be realized by creating technical and communicational solutions for the difficulties outlined above. Our design focuses on the interaction of the different roles within the scientific research and publication culture. The executable research compendium reduces efforts on the technical side of reproducibility and thus fosters the community’s acceptance of openness and reproducibility and creates the basis for open collaborations between scientists.
6 Future Work
Because ERCs only work when taking care of the specifics of both a scientific domain and the software, we expect a focussed solution to deliver best results for users. We are currently in the process of developing an open, formal specification for ERCs and an open source web-platform allowing users to build, store, execute, and interact with ERCs in the context of computational geosciences in R, going beyond most current research data platforms (see Opening Reproducible Research project on GitHub).
The prototypical implementation will be subject to a series of usability evaluations considering the views of all stakeholders and roles. In particular, the usability while creating ERCs will be a crucial factor for the acceptance of ERCs as a form of publication. Although ERCs are designed with existing platforms and workflows in mind, a practical evaluation of their successful integration is needed, i.e. a demonstration of a complete publishing process from submission on a journal platform, evaluation during review, storage in repositories, publication and interaction on an online platform, and long-term archival. Inherently manual steps of a publishing workflow, e.g. copy-editing, create new challenges for systematic interpretation of the output. A user friendly interactive execution of compendia has to address open questions regarding parameter transfer and partial container execution.
Acknowledgements
This work is supported by the project Opening Reproducible Research (see also Offene Reproduzierbare Forschung) funded by the German Research Foundation (DFG) under project numbers PE 1632/10-1, KR 3930/3-1 and TR 864/6-1.
References
| [1] |
Barnes, Nick. "Publish Your Computer Code: It Is Good Enough." Nature News 467, no. 7317 (October 13, 2010): 753-753. https://doi.org/10.1038/467753a |
| [2] |
Bechhofer, Sean, Iain Buchan, David De Roure, Paolo Missier, John Ainsworth, Jiten Bhagat, Philip Couch, et al. "Why Linked Data Is Not Enough for Scientists". Future Generation Computer Systems, Special section: Recent advances in e-Science 29, no. 2 (February 2013): 599-611. https://doi.org/10.1016/j.future.2011.08.004 |
| [3] |
Belhajjame, Khalid, Jun Zhao, Daniel Garijo, Matthew Gamble, Kristina Hettne, Raul Palma, Eleni Mina, et al. "Using a Suite of Ontologies for Preserving Workflow-Centric Research Objects". Web Semantics: Science, Services and Agents on the World Wide Web 32 (May 2015): 16-42. https://doi.org/10.1016/j.websem.2015.01.003 |
| [4] |
Bizer, Christian, Tom Heath, and Tim Berners-Lee. "Linked Data - The Story So Far." International Journal on Semantic Web and Information Systems 5, no. 3 (33 2009): 1-22. https://doi.org/10.4018/jswis.2009081901 |
| [5] |
Bloom, T., Ganley, E., & Winker, M. (2014). Data access for the open access literature: PLOS's data policy. PLoS Biol 12(2), https://doi.org/10.1371/journal.pbio.1001797 |
| [6] |
Boettiger, C. (2015). An introduction to Docker for reproducible research. ACM SIGOPS Operating Systems Review 49(1), 71-79. https://doi.org/10.1145/2723872.2723882 |
| [7] |
Bollen, K., Cacioppo, J. T., Kaplan, R. M., Krosnick, J. A., & Olds, J. L. (2015). Social, behavioral, and economic sciences perspectives on robust and reliable science: Report of the Subcommittee on Replicability in Science, Advisory Committee to the National Science Foundation Directorate for Social, Behavioral, and Economic Sciences. |
| [8] |
Borgman, C. (2007). Scholarship in the digital age: information, infrastructure and the internet. MIT University Press Group Ltd., p. 336. ISBN: 9780262026192 |
| [9] |
Brase, J. "DataCite — A Global Registration Agency for Research Data." In Fourth International Conference on Cooperation and Promotion of Information Resources in Science and Technology, 2009. COINFO '09, 257-61, 2009. https://doi.org/10.1109/COINFO.2009.66 |
| [10] |
Buckheit, Jonathan B., and David L. Donoho. "WaveLab and Reproducible Research". In Wavelets and Statistics, edited by Anestis Antoniadis and Georges Oppenheim, 55-81. Lecture Notes in Statistics 103. Springer New York, 1995. https://doi.org/10.1007/978-1-4612-2544-7_5 |
| [11] |
CCSDS (2012). Consultative Committee for Space Data Systems, Reference model for an open archival information system (OAIS), Magenta Book CCSDS 650.0-M-2, Open Archives Initiative, 2012. |
| [12] |
Chen, Xiaoli, Sünje Dallmeier-Tiessen, Anxhela Dani, Robin Dasler, Javier Delgado Fernández, Pamfilos Fokianos, Patricia Herterich, and Tibor Šimko. "CERN Analysis Preservation: A Novel Digital Library Service to Enable Reusable and Reproducible Research". In Research and Advanced Technology for Digital Libraries, edited by Norbert Fuhr, László Kovács, Thomas Risse, and Wolfgang Nejdl, 9819:347-56. Cham: Springer International Publishing, 2016. https://doi.org/10.1007/978-3-319-43997-6_27 |
| [13] |
Chirigati, F., Shasha, D., & Freire, J. (2013). Reprozip: Using provenance to support computational reproducibility. Proceedings of the
5th USENIX Workshop on the Theory and Practice of Provenance, Lombard, IL, USA, April 2-3, 2013. USENIX Association Berkeley, CA, USA.
|
| [14] |
Clarkson, Chris, Mike Smith, Ben Marwick, Richard Fullagar, Lynley A. Wallis, Patrick Faulkner, Tiina Manne, et al. "The Archaeology, Chronology and Stratigraphy of Madjedbebe (Malakunanja II): A Site in Northern Australia with Early Occupation". Journal of Human Evolution 83 (June 2015): 46-64. https://doi.org/10.1016/j.jhevol.2015.03.014 |
| [15] |
Dua, R., A. R. Raja, and D. Kakadia. "Virtualization vs Containerization to Support PaaS". In 2014 IEEE International Conference on Cloud Engineering (IC2E), 610-14, 2014. https://doi.org/10.1109/IC2E.2014.41 |
| [16] |
Easterbrook, S. M. (2014). Open code for open science?. Nature Geoscience 7(11), 779-781. https://doi.org/10.1038/ngeo2283 |
| [17] |
European Commission (2015). Access to and preservation of scientific information in Europe. Report on the implementation of Commission Recommendation C(2012) 4890 final. https://doi.org/10.2777/975917 |
| [18] |
Gentleman, R., & Temple Lang, D. (2007). Statistical analyses and reproducible research. Journal of Computational and Graphical Statistics 16:2, 1-23, https://doi.org/10.1198/106186007X178663 |
| [19] |
Glandon, P. (2011). "Appendix to the Report of the Editor: Report on the American Economic Review Data Availability Compliance Project. American Economic Review 101(3): 695-699. |
| [20] |
Goecks, Jeremy, Anton Nekrutenko, and James Taylor. "Galaxy: A Comprehensive Approach for Supporting Accessible, Reproducible, and Transparent Computational Research in the Life Sciences". Genome Biology 11 (2010): R86. https://doi.org/10.1186/gb-2010-11-8-r86 |
| [21] |
Goodman, S. N., Fanelli, D., & Ioannidis, J. P. (2016). What does research reproducibility mean? Science Translational Medicine 8(341), 341ps12-341ps12. https://doi.org/10.1126/scitranslmed.aaf5027 |
| [22] |
Haak, Laurel L., Martin Fenner, Laura Paglione, Ed Pentz, and Howard Ratner. "ORCID: A System to Uniquely Identify Researchers." Learned Publishing 25, no. 4 (October 1, 2012): 259-64. https://doi.org/10.1087/20120404 |
| [23] |
Hanson, Karen L., Tim DiLauro, and Mark Donoghue. "The RMap Project: Capturing and Preserving Associations Amongst Multi-Part Distributed Publications". In Proceedings of the 15th ACM/IEEE-CS Joint Conference on Digital Libraries, 281-282. JCDL '15. New York, NY, USA: ACM, 2015. https://doi.org/10.1145/2756406.2756952 |
| [24] |
Harnad, Stevan, et al. "The access/impact problem and the green and gold roads to open access." Serials Review 30.4 (2004): 310-314. https://doi.org/10.1080/00987913.2004.10764930 |
| [25] |
Herndon, T., Ash, M., & Pollin, R. (2014). Does high public debt consistently stifle economic growth? A critique of Reinhart and Rogoff. Cambridge Journal of Economics, 38(2), 257-279. https://doi.org/10.1093/cje/bet075 |
| [26] |
Howe, B. (2012). Virtual appliances, cloud computing, and reproducible research. Computing in Science & Engineering, 14(4), 36-41. https://doi.org/10.1109/MCSE.2012.62 |
| [27] |
Hung, L. H., Kristiyanto, D., Lee, S. B., & Yeung, K. Y. (2016). GUIdock: Using Docker Containers with a Common Graphics User Interface to Address the Reproducibility of Research. PloS one, 11(4), e0152686. |
| [28] |
Knuth, Donald E. "Literate Programming." The Computer Journal 27, no. 2 (May 1984): 97-111. https://doi.org/10.1093/comjnl/27.2.97 |
| [29] |
Kunze, J., Littman, J., Madden L., Summers, E., Boyko, A. & Vargas, B. "The bagit file packaging format (v0. 97)." Washington DC (2011). |
| [30] |
Leek, J. T., & Peng, R. D. (2015). Opinion: Reproducible research can still be wrong: Adopting a prevention approach. Proceedings of the National Academy of Sciences, 112(6), 1645-1646. |
| [31] |
Markowetz, Florian. "Five Selfish Reasons to Work Reproducibly." Genome Biology 16 (2015): 274. https://doi.org/10.1186/s13059-015-0850-7 |
| [32] |
Marwick, Ben. "1989-Excavation-Report-Madjebebe." March 23, 2015. https://doi.org/10.6084/m9.figshare.1297059.v2 |
| [33] |
Mecklenburg, R. (2004). Managing Projects with GNU Make, 3rd Edition. O'Reilly Media, ISBN: 0-596-00610-1. |
| [34] |
Merkel, D. (2014). Docker: lightweight linux containers for consistent development and deployment. Linux Journal, 2014(239), 2, ISSN: 1075-3583. |
| [35] |
Nature Editorial. "Code Share". Nature 514, no. 7524 (2014): 536-536. https://doi.org/10.1038/514536a |
| [36] |
Nosek, B. A., G. Alter, G. C. Banks, D. Borsboom, S. D. Bowman, S. J. Breckler, S. Buck, et al. "Promoting an Open Research Culture". Science 348, no. 6242 (26 June 2015): 1422-25. https://doi.org/10.1126/science.aab2374 |
| [37] |
Palma, Raúl, Oscar Corcho, Piotr Hołubowicz, Sara Pérez, Kevin Page, and Cezary Mazurek. "Digital Libraries for the Preservation of Research Methods and Associated Artifacts". In Proceedings of the 1st International Workshop on Digital Preservation of Research Methods and Artefacts, 8-15. DPRMA '13. New York, NY, USA: ACM, 2013. https://doi.org/10.1145/2499583.2499589 |
| [38] |
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2014. |
| [39] |
Reinhart, C. M., & Rogoff, K. S. (2010). Growth in a time of debt (digest summary). American Economic Review, 100(2), 573-578. https://doi.org/10.2469/dig.v40.n3.19 |
| [40] |
Santana-Perez, I., Ferreira da Silva, R., Rynge, M., Deelman, E., Pérez-Hernández, M. S., Corcho, O. (2017). Reproducibility of Execution Environments in Computational Science Using Semantics and Clouds. Future Generation Computer Systems. https://doi.org/10.1016/j.future.2015.12.017 |
| [41] |
Stodden, V., Leisch, F., & Peng, R. D. (Eds.). (2014). Implementing reproducible research. CRC Press. |
| [42] |
Stodden, V., Miguez, S. and Seiler, J. (2015). Researchcompendia. org: Cyberinfrastructure for reproducibility and collaboration in computational science. Computing in Science & Engineering, 17(1), pp.12-19. |
| [43] |
Thain, D., Ivie P., Meng, H. (2015). Techniques for Preserving Scientific Software Executions: Preserve the Mess or Encourage Cleanliness?. https://doi.org/10.7274/R0CZ353M |
| [44] |
Vandewalle, P., Kovacevic, J., & Vetterli, M. (2009). Reproducible research in signal processing. IEEE Signal Processing Magazine, 26(3), 37-47. https://doi.org/10.1109/MSP.2009.932122 |
| [45] |
Vines, T. H., Albert, A. Y., Andrew, R. L., Débarre, F., Bock, D. G., Franklin, M. T., Rennison, D. J. (2014). The availability of research data declines rapidly with article age. Current Biology 24(1), 94-97. https://doi.org/10.1016/j.cub.2013.11.014 |
| [46] |
Wolstencroft, K., Haines, R., Fellows, D., Williams, A., Withers, D., Owen, S., Soiland-Reyes, S., Dunlop, I., Nenadic, A., Fisher, P., Bhagat, J., Belhajjame, K., Bacall, F., Hardisty, A., Hidalga, A., Vargas, M., Sufi, S., Goble, C. The Taverna Workflow Suite: Designing and Executing Workflows of Web Services on the Desktop, Web or in the Cloud. Nucleic Acids Research 41, no. W1 (1 July 2013): W557-61. https://doi.org/10.1093/nar/gkt328 |
About the Authors
Daniel Nüst is a researcher at the Institute for Geoinformatics, University of Münster. He completed his studies in Münster with a Diploma in Geoinformatics in 2011 and afterwards worked at 52°North Initiative for Geospatial Open Source Software as a consultant and software developer. Since 2016 he has worked on creating, storing and executing reproducible research packages in the DFG-project "Opening Reproducible Research".
Markus Konkol is a research associate in the DFG-project "Opening Reproducible Research" at the Institute for Geoinformatics, University of Münster. He is currently doing his Ph.D. in the DFG-funded project Opening Reproducible Research that aims at facilitating access to and interaction with research results. His main topic is about interconnecting the textual publication, data sets, source code and UI elements in order to assist scientists in exploring and creating dynamic publications.
Marc Schutzeichel is a research associate in the DFG-project "Opening Reproducible Research". He works at the University and State Library, Münster.
Edzer Pebesma is professor of Geoinformatics at University of Münster since 2007. He is developer and maintainer of several popular R packages for handling and analyzing spatial and spatiotemporal data (sp, spacetime, gstat), co-author of the book Applied Spatial Data Analysis with R, and active member of the r-sig-geo community. He is Co-Editor-in-Chief for the Journal of Statistical Software and Computers & Geosciences, and associate editor for Spatial Statistics.
Chris Kray is a professor of Geoinformatics at the Institute for Geoinformatics (ifgi) at the University of Münster. His research interests include location-based services, smart cities and human-computer interaction (in particular: interaction with spatial information). Chris is the scientific coordinator of the ITN "GEO-C: enabling open cities" at ifgi, where he works on realising transparency, accessibility and privacy protection in the context of smart cities.
Holger Przibytzin is Department Manager of Scientific Information Systems at the University and State Library, Münster.
Jörg Lorenz is Head of the Science and Innovation department of the University and State Library, Münster.